You are browsing environment: FUNGIDB
CAZyme Information: PWY67369.1
You are here: Home > Sequence: PWY67369.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
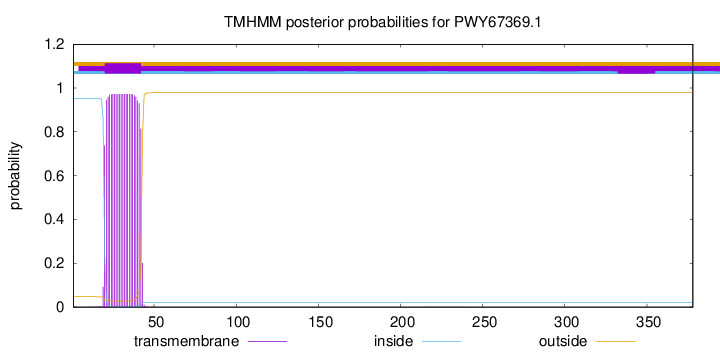
TMHMM annotations
Basic Information help
| Species | Aspergillus eucalypticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus eucalypticola | |||||||||||
| CAZyme ID | PWY67369.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | SGNH_hydro domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A317V124] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 120 | 347 | 4.6e-50 | 0.979381443298969 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 2.11e-39 | 198 | 350 | 14 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 2.22e-07 | 217 | 342 | 54 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238141 | SGNH_hydrolase | 5.60e-06 | 210 | 346 | 51 | 184 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 238868 | XynE_like | 1.29e-04 | 218 | 350 | 68 | 204 | SGNH_hydrolase subfamily, similar to the putative arylesterase/acylhydrolase from the rumen anaerobe Prevotella bryantii XynE. The P. bryantii XynE gene is located in a xylanase gene cluster. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 238872 | SGNH_hydrolase_like_2 | 1.43e-04 | 204 | 347 | 46 | 189 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.81e-236 | 1 | 378 | 23 | 395 | |
| 3.96e-119 | 94 | 371 | 32 | 311 | |
| 2.86e-92 | 165 | 371 | 19 | 222 | |
| 4.17e-90 | 120 | 371 | 512 | 769 | |
| 3.96e-85 | 116 | 371 | 332 | 591 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
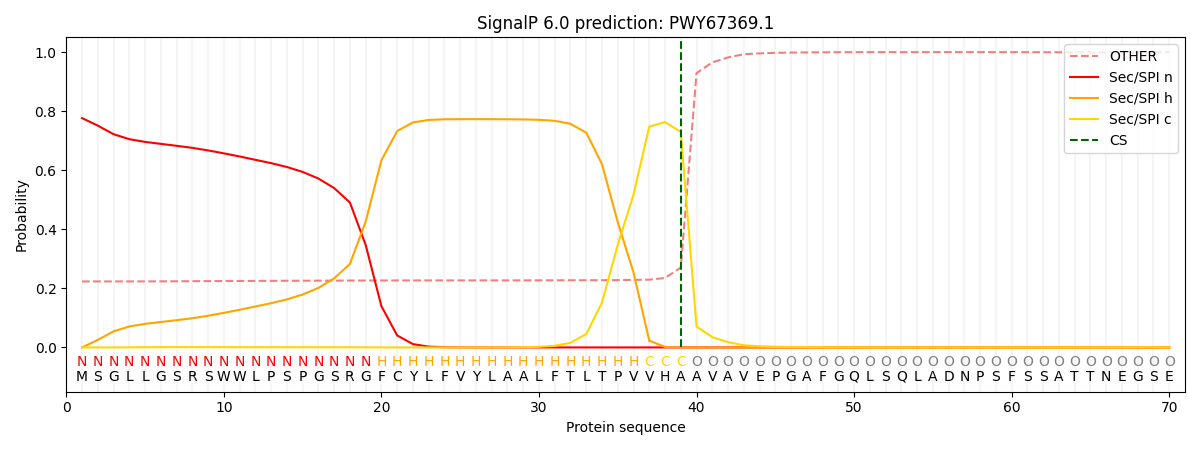
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.239804 | 0.760182 | CS pos: 39-40. Pr: 0.7295 |