You are browsing environment: FUNGIDB
CAZyme Information: PWY61885.1
You are here: Home > Sequence: PWY61885.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus eucalypticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus eucalypticola | |||||||||||
| CAZyme ID | PWY61885.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | cutinase-domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 118 | 280 | 7.3e-46 | 0.9841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 1.99e-48 | 118 | 281 | 1 | 172 | Cutinase. |
| 223477 | YpfH | 9.64e-07 | 180 | 284 | 82 | 177 | Predicted esterase [General function prediction only]. |
| 369775 | PE-PPE | 0.003 | 155 | 230 | 2 | 89 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
| 396693 | Abhydrolase_2 | 0.005 | 193 | 264 | 103 | 179 | Phospholipase/Carboxylesterase. This family consists of both phospholipases and carboxylesterases with broad substrate specificity, and is structurally related to alpha/beta hydrolases pfam00561. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.56e-128 | 1 | 286 | 1 | 288 | |
| 2.55e-124 | 1 | 286 | 1 | 291 | |
| 2.55e-124 | 1 | 286 | 1 | 291 | |
| 6.79e-124 | 1 | 286 | 1 | 289 | |
| 9.41e-86 | 109 | 284 | 36 | 210 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.68e-36 | 119 | 279 | 79 | 244 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 4.77e-27 | 115 | 282 | 29 | 209 | Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005],5AJH_B Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005],5AJH_C Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005] |
|
| 6.51e-27 | 117 | 283 | 19 | 197 | Crystal structure of Aspergillus oryzae cutinase [Aspergillus oryzae] |
|
| 1.93e-26 | 112 | 280 | 24 | 205 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 5.34e-26 | 117 | 279 | 10 | 184 | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon [Aspergillus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.95e-75 | 108 | 284 | 50 | 223 | Cutinase 4 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut4 PE=2 SV=1 |
|
| 3.01e-38 | 118 | 284 | 30 | 202 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 8.19e-38 | 118 | 283 | 30 | 200 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 6.22e-36 | 118 | 278 | 78 | 243 | Cutinase cut1 OS=Trichoderma harzianum OX=5544 GN=cut1 PE=1 SV=1 |
|
| 3.40e-35 | 119 | 279 | 79 | 244 | Cutinase OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=M419DRAFT_76732 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
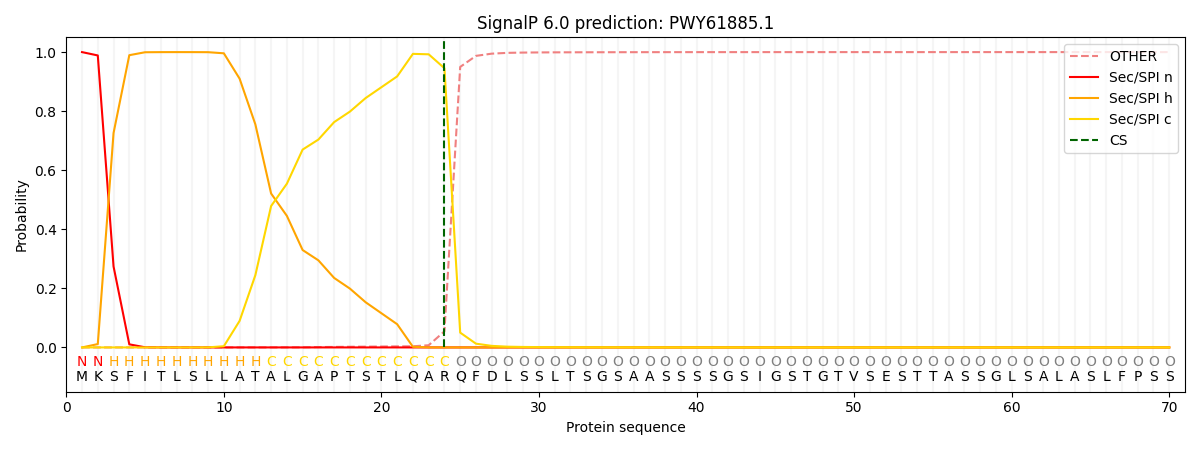
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000338 | 0.999634 | CS pos: 24-25. Pr: 0.9455 |
