You are browsing environment: FUNGIDB
CAZyme Information: PV10_06874-t45_1-p1
You are here: Home > Sequence: PV10_06874-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala mesophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala mesophila | |||||||||||
| CAZyme ID | PV10_06874-t45_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 33 | 452 | 1.9e-114 | 0.995049504950495 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 1.99e-121 | 34 | 452 | 1 | 383 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.79e-102 | 6 | 480 | 2 | 590 | |
| 2.27e-101 | 26 | 454 | 38 | 533 | |
| 3.14e-100 | 21 | 455 | 22 | 571 | |
| 3.48e-100 | 26 | 454 | 38 | 533 | |
| 9.70e-100 | 26 | 454 | 38 | 533 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.43e-97 | 26 | 471 | 27 | 560 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=alg10 PE=3 SV=1 |
|
| 9.31e-94 | 26 | 457 | 60 | 590 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=ALG10 PE=3 SV=1 |
|
| 2.47e-92 | 26 | 455 | 28 | 547 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg10 PE=3 SV=1 |
|
| 3.11e-73 | 21 | 474 | 62 | 654 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ALG10 PE=3 SV=1 |
|
| 5.24e-73 | 25 | 463 | 60 | 676 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-10 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000028 | 0.000003 |
TMHMM Annotations download full data without filtering help
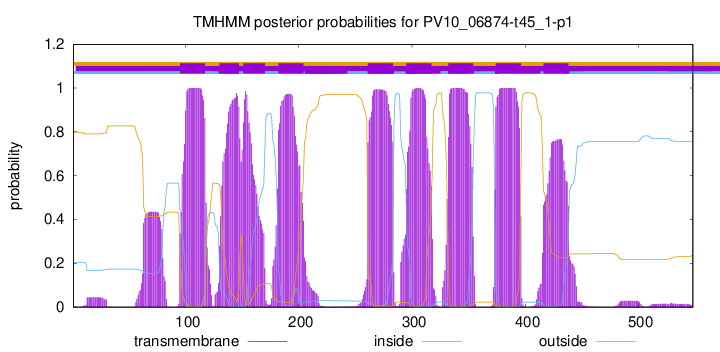
| Start | End |
|---|---|
| 95 | 117 |
| 130 | 147 |
| 151 | 170 |
| 182 | 204 |
| 261 | 283 |
| 295 | 317 |
| 332 | 354 |
| 374 | 396 |
| 416 | 438 |
