You are browsing environment: FUNGIDB
CAZyme Information: PV10_03614-t45_1-p1
You are here: Home > Sequence: PV10_03614-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala mesophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala mesophila | |||||||||||
| CAZyme ID | PV10_03614-t45_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 64 | 475 | 1.1e-58 | 0.8296089385474861 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 4.60e-21 | 197 | 481 | 116 | 339 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 8.19e-05 | 200 | 411 | 119 | 276 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.24e-193 | 44 | 542 | 70 | 569 | |
| 6.00e-186 | 51 | 542 | 55 | 599 | |
| 9.56e-186 | 51 | 542 | 69 | 613 | |
| 9.56e-186 | 51 | 542 | 69 | 613 | |
| 1.69e-185 | 44 | 542 | 49 | 548 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.64e-07 | 179 | 360 | 96 | 239 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 5.71e-07 | 179 | 360 | 99 | 242 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 6.09e-07 | 179 | 360 | 117 | 260 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 6.09e-07 | 179 | 360 | 117 | 260 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5AGD_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBM_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
|
| 8.96e-07 | 179 | 360 | 130 | 273 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.91e-06 | 191 | 409 | 138 | 315 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
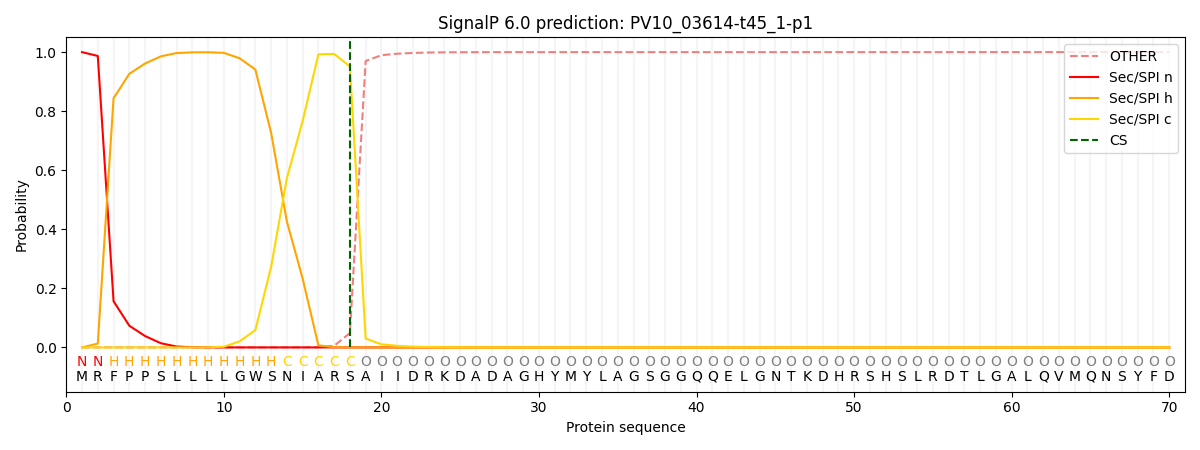
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000710 | 0.999250 | CS pos: 18-19. Pr: 0.9513 |
