You are browsing environment: FUNGIDB
CAZyme Information: PV10_02410-t45_1-p1
You are here: Home > Sequence: PV10_02410-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala mesophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala mesophila | |||||||||||
| CAZyme ID | PV10_02410-t45_1-p1 | |||||||||||
| CAZy Family | AA8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 30 | 426 | 9.2e-156 | 0.9850374064837906 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 1.42e-22 | 1 | 428 | 14 | 396 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.09e-140 | 12 | 428 | 10 | 426 | |
| 4.98e-133 | 13 | 428 | 12 | 431 | |
| 1.54e-132 | 30 | 435 | 35 | 434 | |
| 4.37e-132 | 30 | 435 | 35 | 434 | |
| 7.78e-130 | 5 | 434 | 7 | 440 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.47e-112 | 36 | 426 | 11 | 400 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.52e-112 | 36 | 426 | 12 | 401 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.57e-112 | 36 | 426 | 13 | 402 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.11e-79 | 26 | 428 | 5 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.02e-12 | 36 | 427 | 72 | 442 | L-sorbosone dehydrogenase OS=Gluconacetobacter liquefaciens OX=89584 PE=4 SV=1 |
|
| 3.96e-11 | 38 | 426 | 35 | 374 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
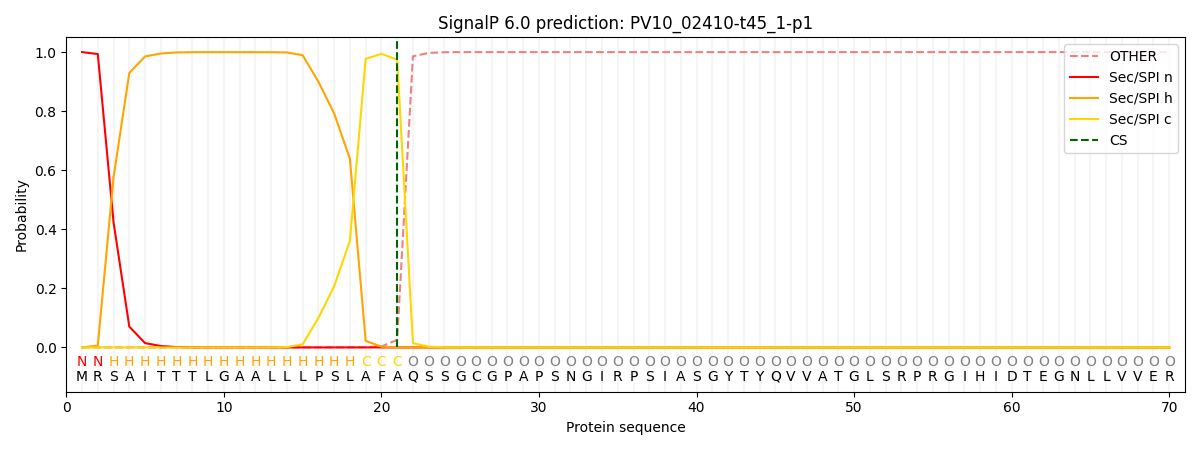
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000403 | 0.999564 | CS pos: 21-22. Pr: 0.9743 |
