You are browsing environment: FUNGIDB
CAZyme Information: PV07_05165-t44_1-p1
You are here: Home > Sequence: PV07_05165-t44_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cladophialophora immunda | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora immunda | |||||||||||
| CAZyme ID | PV07_05165-t44_1-p1 | |||||||||||
| CAZy Family | GH15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.59:2 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397634 | Glyco_hydro_71 | 7.68e-155 | 23 | 402 | 1 | 369 | Glycosyl hydrolase family 71. Family of alpha-1,3-glucanases. |
| 211418 | GH71 | 4.56e-114 | 20 | 297 | 4 | 282 | Glycoside hydrolase family 71. This family of glycoside hydrolases 71 (following the CAZY nomenclature) function as alpha-1,3-glucanases (mutanases, EC 3.2.1.59). They appear to have an endo-hydrolytic mode of enzymatic activity and bacterial members are investigated as candidates for the development of dental caries treatments.The member from fission yeast, endo-alpha-1,3-glucanase Agn1p, plays a vital role in daughter cell separation, while Agn2p has been associated with endolysis of the ascus wall. |
| 178203 | PLN02593 | 8.98e-58 | 516 | 620 | 3 | 107 | adrenodoxin-like ferredoxin protein |
| 211414 | GH99_GH71_like | 1.03e-43 | 75 | 293 | 48 | 284 | Glycoside hydrolase families 71, 99, and related domains. This superfamily of glycoside hydrolases contains families GH71 and GH99 (following the CAZY nomenclature), as well as other members with undefined function and specificity. |
| 131062 | fdx_isc | 3.60e-28 | 527 | 619 | 17 | 105 | ferredoxin, 2Fe-2S type, ISC system. This family consists of proteobacterial ferredoxins associated with and essential to the ISC system of 2Fe-2S cluster assembly. This family is closely related to (but excludes) eukaryotic (mitochondrial) adrenodoxins, which are ferredoxins involved in electron transfer to P450 cytochromes. [Biosynthesis of cofactors, prosthetic groups, and carriers, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.79e-199 | 9 | 429 | 8 | 432 | |
| 9.35e-136 | 15 | 428 | 48 | 467 | |
| 9.35e-136 | 15 | 428 | 48 | 467 | |
| 1.04e-126 | 29 | 428 | 16 | 417 | |
| 1.36e-126 | 20 | 428 | 20 | 431 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.70e-49 | 511 | 625 | 1 | 114 | Oxidized Yeast Adrenodoxin Homolog 1 [Saccharomyces cerevisiae S288C],2MJE_A Reduced Yeast Adrenodoxin Homolog 1 [Saccharomyces cerevisiae S288C] |
|
| 2.29e-31 | 516 | 620 | 4 | 110 | Backbone 1H, 13C, and 15N Chemical Shift Assignments and structure of Iron-sulfur cluster binding protein from Ehrlichia chaffeensis [Ehrlichia chaffeensis str. Arkansas] |
|
| 1.42e-30 | 511 | 614 | 1 | 103 | Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria [Schizosaccharomyces pombe],2WLB_B Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria [Schizosaccharomyces pombe] |
|
| 2.03e-25 | 512 | 623 | 4 | 116 | Crystal structure of human CYP11A1 in complex with 22-hydroxycholesterol [Homo sapiens],3N9Z_D Crystal structure of human CYP11A1 in complex with 22-hydroxycholesterol [Homo sapiens],3NA1_C Crystal structure of human CYP11A1 in complex with 20-hydroxycholesterol [Homo sapiens],3NA1_D Crystal structure of human CYP11A1 in complex with 20-hydroxycholesterol [Homo sapiens] |
|
| 2.62e-25 | 512 | 623 | 6 | 118 | Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_B Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_C Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_D Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_E Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_F Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_G Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens],3P1M_H Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.88e-65 | 21 | 428 | 22 | 420 | Glucan endo-1,3-alpha-glucosidase agn1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn1 PE=1 SV=2 |
|
| 5.04e-58 | 23 | 428 | 38 | 428 | Mutanase Pc12g07500 OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=PCH_Pc12g07500 PE=1 SV=1 |
|
| 6.59e-56 | 494 | 625 | 41 | 171 | Adrenodoxin homolog, mitochondrial OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YAH1 PE=1 SV=1 |
|
| 9.89e-35 | 21 | 321 | 11 | 329 | Ascus wall endo-1,3-alpha-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn2 PE=1 SV=2 |
|
| 2.64e-34 | 512 | 625 | 79 | 192 | Adrenodoxin-like protein 2, mitochondrial OS=Arabidopsis thaliana OX=3702 GN=MFDX2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
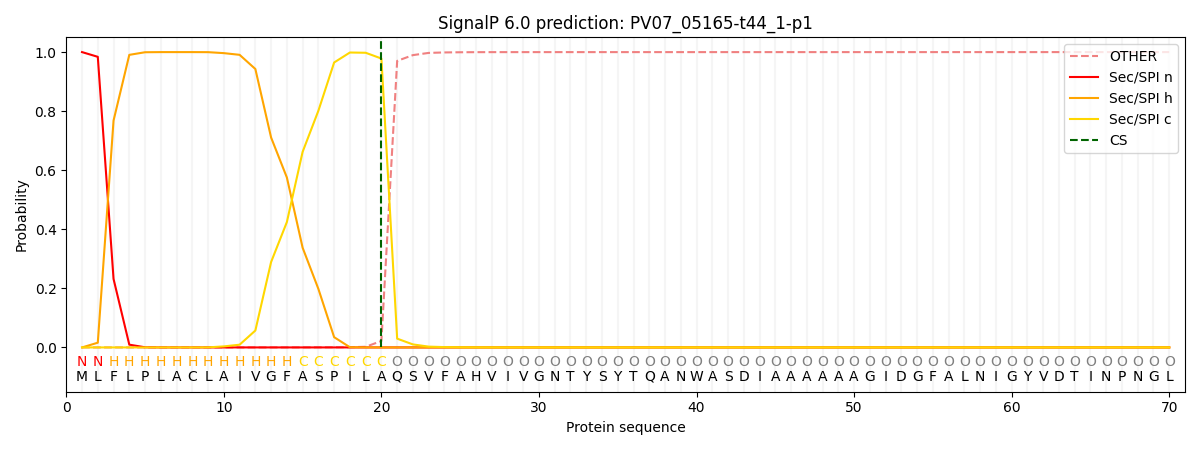
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000242 | 0.999736 | CS pos: 20-21. Pr: 0.9782 |
