You are browsing environment: FUNGIDB
CAZyme Information: PV06_10418-t43_1-p1
You are here: Home > Sequence: PV06_10418-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
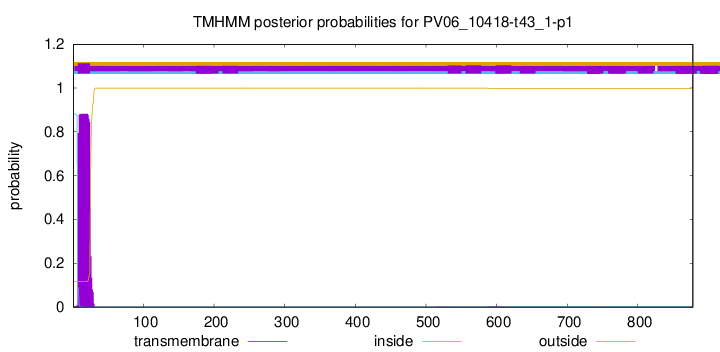
TMHMM annotations
Basic Information help
| Species | Exophiala oligosperma | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala oligosperma | |||||||||||
| CAZyme ID | PV06_10418-t43_1-p1 | |||||||||||
| CAZy Family | GT34 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 199 | 874 | 6.3e-175 | 0.9977578475336323 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 0.0 | 199 | 874 | 1 | 453 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 2.20e-97 | 164 | 875 | 35 | 519 | glycoside hydrolase family 47 protein; Provisional |
| 397265 | 4HBT | 0.003 | 228 | 290 | 5 | 68 | Thioesterase superfamily. This family contains a wide variety of enzymes, principally thioesterases. This family includes 4HBT (EC 3.1.2.23) which catalyzes the final step in the biosynthesis of 4-hydroxybenzoate from 4-chlorobenzoate in the soil dwelling microbe Pseudomonas CBS-3. This family includes various cytosolic long-chain acyl-CoA thioester hydrolases. Long-chain acyl-CoA hydrolases hydrolyze palmitoyl-CoA to CoA and palmitate, they also catalyze the hydrolysis of other long chain fatty acyl-CoA thioesters. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.32e-234 | 121 | 877 | 144 | 899 | |
| 9.54e-231 | 121 | 875 | 136 | 861 | |
| 2.45e-230 | 121 | 875 | 132 | 858 | |
| 4.89e-230 | 121 | 875 | 132 | 858 | |
| 1.55e-225 | 104 | 875 | 113 | 869 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-42 | 192 | 871 | 5 | 448 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 1.70e-42 | 192 | 871 | 10 | 453 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 1.76e-42 | 192 | 871 | 10 | 453 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 6.66e-42 | 192 | 871 | 88 | 531 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
|
| 7.31e-42 | 192 | 871 | 5 | 448 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.85e-46 | 192 | 873 | 98 | 536 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS2 OS=Arabidopsis thaliana OX=3702 GN=MNS2 PE=1 SV=1 |
|
| 2.06e-46 | 181 | 873 | 87 | 535 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 OS=Arabidopsis thaliana OX=3702 GN=MNS1 PE=1 SV=1 |
|
| 2.63e-43 | 190 | 871 | 178 | 623 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Mus musculus OX=10090 GN=Man1a2 PE=1 SV=1 |
|
| 3.54e-43 | 188 | 871 | 176 | 623 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Homo sapiens OX=9606 GN=MAN1A2 PE=1 SV=1 |
|
| 3.54e-42 | 155 | 873 | 9 | 506 | Probable mannosyl-oligosaccharide alpha-1,2-mannosidase ARB_00035 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00035 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000006 | 0.000025 |