You are browsing environment: FUNGIDB
CAZyme Information: PV06_04207-t43_1-p1
Basic Information
help
| Species |
Exophiala oligosperma
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala oligosperma
|
| CAZyme ID |
PV06_04207-t43_1-p1
|
| CAZy Family |
GH13 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1071 |
|
115594.92 |
4.3078 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_EoligospermaCBS72588 |
11938 |
N/A |
45 |
11893
|
|
| Gene Location |
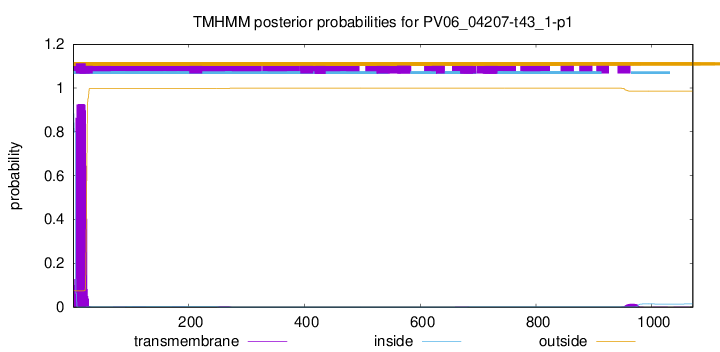
No EC number prediction in PV06_04207-t43_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
299 |
824 |
5.9e-57 |
0.6237864077669902 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 407266
|
Glyco_hydro_106 |
6.54e-14 |
332 |
796 |
427 |
873 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.96e-304 |
27 |
1067 |
19 |
1012 |
| 2.44e-302 |
28 |
1067 |
21 |
994 |
| 4.76e-302 |
29 |
1067 |
25 |
1016 |
| 3.49e-295 |
37 |
1067 |
31 |
1001 |
| 3.49e-295 |
37 |
1067 |
31 |
1001 |
PV06_04207-t43_1-p1 has no PDB hit.
PV06_04207-t43_1-p1 has no Swissprot hit.
This protein is predicted as SP
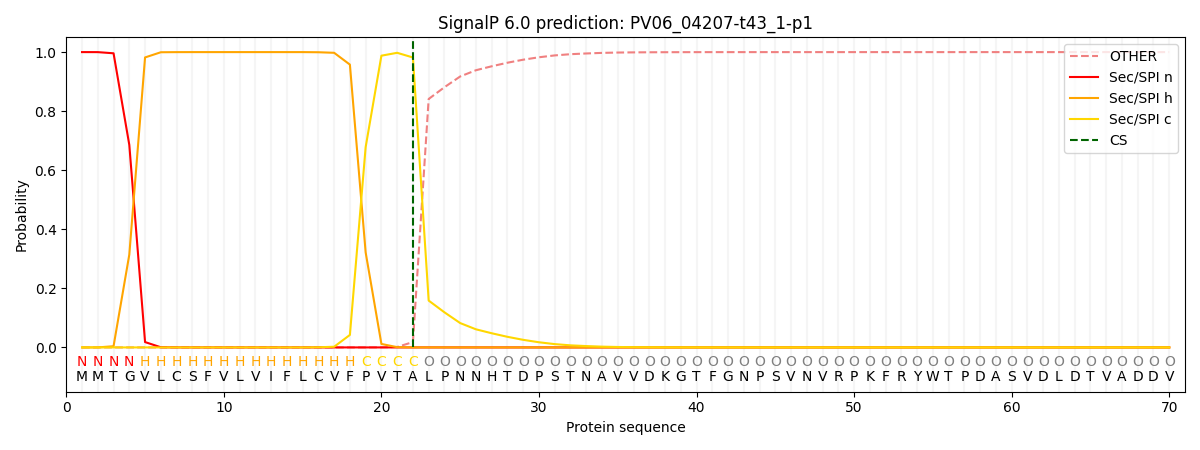
| Other |
SP_Sec_SPI |
CS Position |
| 0.000214 |
0.999761 |
CS pos: 22-23. Pr: 0.9817 |