You are browsing environment: FUNGIDB
PV06_03696-t43_1-p1
Basic Information
help
Species
Exophiala oligosperma
Lineage
Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala oligosperma
CAZyme ID
PV06_03696-t43_1-p1
CAZy Family
GH128
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
339
38785.55
6.5068
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_EoligospermaCBS72588
11938
N/A
45
11893
Gene Location
No EC number prediction in PV06_03696-t43_1-p1.
Family
Start
End
Evalue
family coverage
GT34
78
266
1.4e-22
0.8292682926829268
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
368535
Glyco_transf_34
5.62e-10
76
262
40
231
galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34.
more
215617
PLN03182
1.47e-05
76
201
163
300
xyloglucan 6-xylosyltransferase; Provisional
more
215616
PLN03181
0.001
82
180
170
265
glycosyltransferase; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.28e-68
41
318
78
349
4.29e-68
49
302
176
415
2.74e-67
41
303
78
336
2.74e-67
41
303
78
336
2.74e-67
41
303
78
336
PV06_03696-t43_1-p1 has no PDB hit.
PV06_03696-t43_1-p1 has no Swissprot hit.
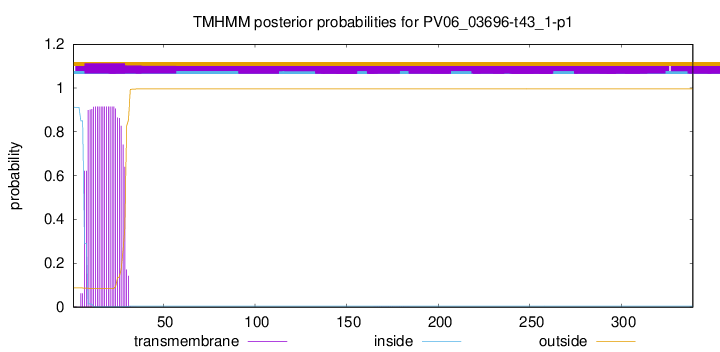
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.998300
0.001711