You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T010362-RA-p1
Basic Information
help
| Species |
Globisporangium ultimum
|
| Lineage |
Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum
|
| CAZyme ID |
PUG3_T010362-RA-p1
|
| CAZy Family |
GH72 |
| CAZyme Description |
Endo-beta-1,6-galactanase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 474 |
|
52179.81 |
7.7950 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_GultimumBR650 |
14086 |
1223559 |
0 |
14086
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH30 |
158 |
467 |
3e-83 |
0.6907894736842105 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227807
|
XynC |
4.78e-10 |
84 |
467 |
78 |
426 |
O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 405300
|
Glyco_hydr_30_2 |
3.60e-07 |
44 |
347 |
18 |
367 |
O-Glycosyl hydrolase family 30. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.41e-181 |
18 |
470 |
21 |
497 |
| 1.19e-149 |
15 |
472 |
22 |
497 |
| 4.94e-149 |
13 |
472 |
22 |
498 |
| 9.12e-147 |
13 |
472 |
22 |
498 |
| 9.12e-147 |
13 |
472 |
22 |
498 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.37e-08 |
141 |
467 |
88 |
384 |
Crystal Structure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.20e-111 |
9 |
474 |
4 |
478 |
Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
This protein is predicted as SP
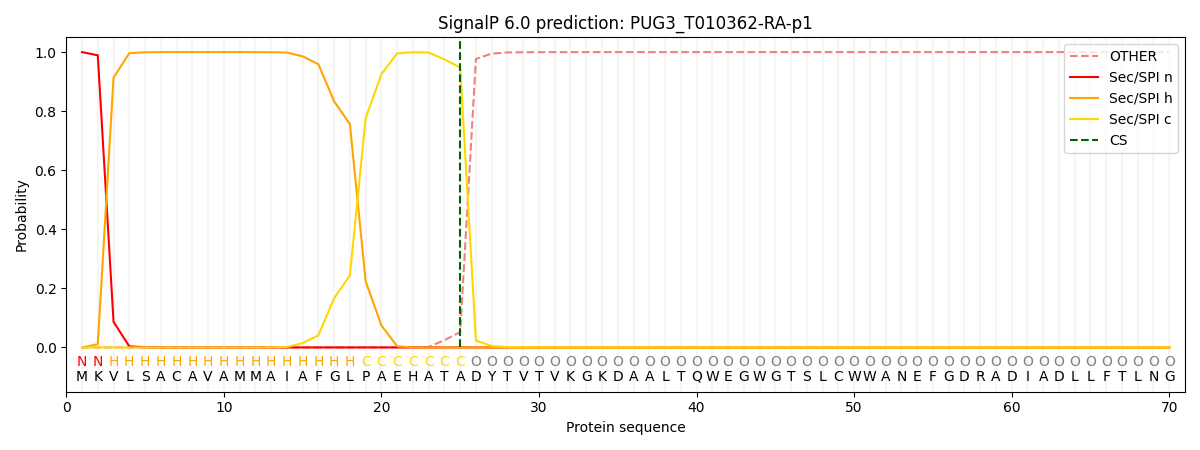
| Other |
SP_Sec_SPI |
CS Position |
| 0.000227 |
0.999761 |
CS pos: 25-26. Pr: 0.9479 |
There is no transmembrane helices in PUG3_T010362-RA-p1.

