You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T010181-RA-p1
You are here: Home > Sequence: PUG3_T010181-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T010181-RA-p1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | Methyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 260 | 874 | 1.8e-56 | 0.5631205673758866 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 7.45e-32 | 242 | 869 | 73 | 609 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 1.06e-18 | 239 | 330 | 4 | 95 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 8.21e-18 | 204 | 297 | 3 | 96 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 223533 | TPR | 1.07e-12 | 72 | 330 | 64 | 290 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
| 276809 | TPR | 2.76e-10 | 270 | 363 | 1 | 85 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.19e-58 | 355 | 873 | 138 | 584 | |
| 7.12e-56 | 363 | 877 | 448 | 887 | |
| 7.12e-56 | 363 | 877 | 448 | 887 | |
| 3.02e-53 | 360 | 886 | 485 | 933 | |
| 5.25e-53 | 355 | 875 | 479 | 922 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-07 | 487 | 850 | 160 | 489 | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_B Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_C Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_D Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
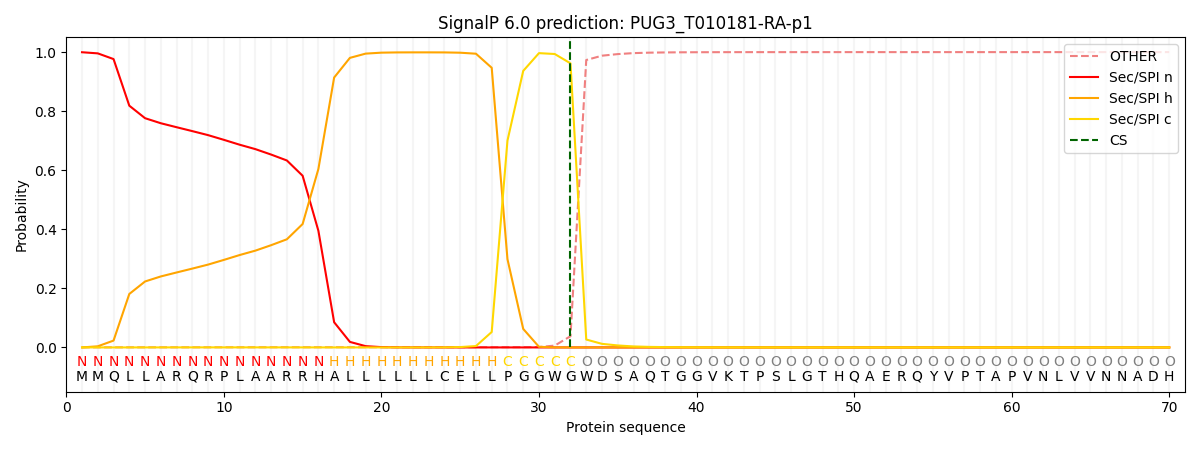
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001146 | 0.998793 | CS pos: 32-33. Pr: 0.9619 |
