You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T003700-RA-p1
You are here: Home > Sequence: PUG3_T003700-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T003700-RA-p1 | |||||||||||
| CAZy Family | GH131 | |||||||||||
| CAZyme Description | Polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 165 | 238 | 2.7e-20 | 0.3711340206185567 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 4.64e-35 | 155 | 333 | 1 | 198 | Pectate lyase. |
| 282710 | PT | 9.89e-07 | 115 | 149 | 2 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| 411100 | internalin_K | 2.72e-06 | 118 | 159 | 509 | 549 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
| 237030 | kgd | 1.79e-04 | 87 | 167 | 40 | 120 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 368653 | Trypan_PARP | 5.30e-04 | 87 | 169 | 51 | 132 | Procyclic acidic repetitive protein (PARP). This family consists of several Trypanosoma brucei procyclic acidic repetitive protein (PARP) like sequences. The procyclic acidic repetitive protein (parp) genes of Trypanosoma brucei encode a small family of abundant surface proteins whose expression is restricted to the procyclic form of the parasite. They are found at two unlinked loci, parpA and parpB; transcription of both loci is developmentally regulated. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.26e-36 | 157 | 342 | 60 | 268 | |
| 1.25e-32 | 153 | 345 | 31 | 250 | |
| 1.30e-31 | 153 | 345 | 31 | 250 | |
| 5.16e-31 | 153 | 345 | 21 | 242 | |
| 2.97e-25 | 155 | 342 | 19 | 236 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.61e-25 | 170 | 342 | 39 | 234 | Probable pectate lyase G OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyG PE=3 SV=1 |
|
| 6.28e-21 | 169 | 333 | 53 | 239 | Probable pectate lyase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyD PE=3 SV=1 |
|
| 3.52e-20 | 159 | 342 | 35 | 241 | Probable pectate lyase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyD PE=3 SV=1 |
|
| 5.51e-20 | 176 | 333 | 57 | 235 | Pectate lyase H OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyH PE=1 SV=1 |
|
| 3.56e-19 | 159 | 329 | 39 | 229 | Probable pectate lyase G OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
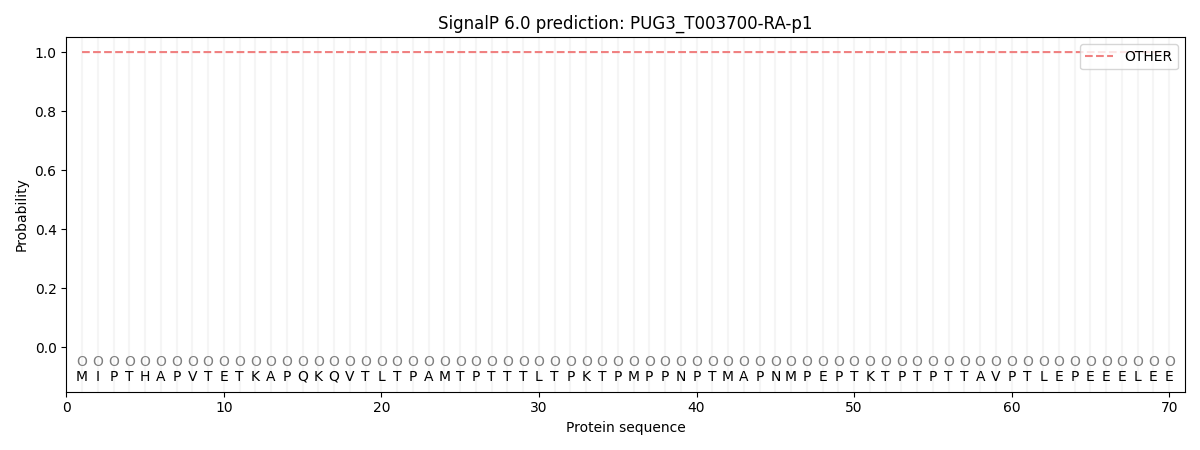
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000079 | 0.000001 |
