You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T003386-RA-p1
You are here: Home > Sequence: PUG3_T003386-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T003386-RA-p1 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | Callose synthase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 436 | 1160 | 1.1e-258 | 0.9228687415426252 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 441 | 1121 | 7 | 700 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1220 | 448 | 1668 | |
| 0.0 | 4 | 1219 | 403 | 1665 | |
| 0.0 | 15 | 1222 | 415 | 1641 | |
| 0.0 | 2 | 1211 | 413 | 1643 | |
| 4.52e-310 | 23 | 1218 | 451 | 1621 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.11e-189 | 8 | 1126 | 634 | 1704 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 6.32e-182 | 21 | 1179 | 663 | 1757 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 1.61e-181 | 2 | 1119 | 631 | 1725 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 3.98e-181 | 23 | 1121 | 647 | 1724 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
|
| 1.46e-180 | 23 | 1121 | 647 | 1724 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999937 | 0.000106 |
TMHMM Annotations download full data without filtering help
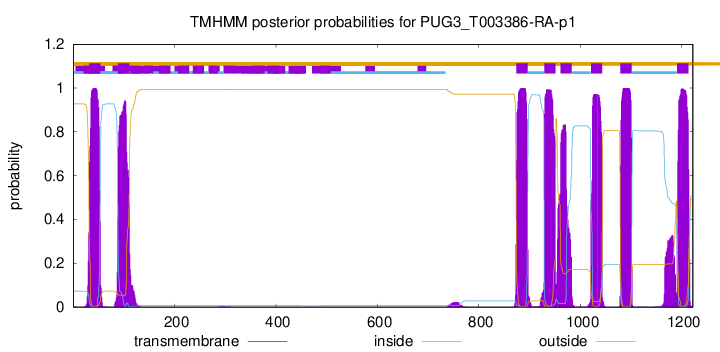
| Start | End |
|---|---|
| 32 | 54 |
| 89 | 111 |
| 874 | 896 |
| 929 | 951 |
| 961 | 983 |
| 1021 | 1043 |
| 1079 | 1101 |
| 1191 | 1213 |
