You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T002373-RA-p1
You are here: Home > Sequence: PUG3_T002373-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T002373-RA-p1 | |||||||||||
| CAZy Family | CBM9 | |||||||||||
| CAZyme Description | Beta-1,4-galactosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223587 | SrmB | 2.50e-38 | 579 | 866 | 180 | 502 | Superfamily II DNA and RNA helicase [Replication, recombination and repair]. |
| 132999 | b4GalT | 1.03e-34 | 55 | 262 | 3 | 219 | Beta-4-Galactosyltransferase is involved in the formation of the poly-N-acetyllactosamine core structures present in glycoproteins and glycosphingolipids. Beta-4-Galactosyltransferase transfers galactose from uridine diphosphogalactose to the terminal beta-N-acetylglucosamine residues, hereby forming the poly-N-acetyllactosamine core structures present in glycoproteins and glycosphingolipids. At least seven homologous beta-4-galactosyltransferase isoforms have been identified that use different types of glycoproteins and glycolipids as substrates. Of the seven identified members of the beta-1,4-galactosyltransferase subfamily (beta1,4-Gal-T1 to -T7), b1,4-Gal-T1 is most characterized (biochemically). It is a Golgi-resident type II membrane enzyme with a cytoplasmic domain, membrane spanning region, and a stem region and catalytic domain facing the lumen. |
| 350715 | DEADc_DDX52 | 4.87e-29 | 579 | 636 | 141 | 198 | DEAD-box helicase domain of DEAD box protein 52. DDX52 (also called ROK1 and HUSSY19) is ubiquitously expressed in testis, endometrium, and other tissues in humans. DDX52 is a member of the DEAD-box helicases, a diverse family of proteins involved in ATP-dependent RNA unwinding, needed in a variety of cellular processes including splicing, ribosome biogenesis and RNA degradation. The name derives from the sequence of the Walker B motif (motif II). This domain contains the ATP-binding region. |
| 350174 | SF2_C_DEAD | 1.38e-23 | 649 | 722 | 31 | 108 | C-terminal helicase domain of the DEAD box helicases. DEAD-box helicases comprise a diverse family of proteins involved in ATP-dependent RNA unwinding, needed in a variety of cellular processes including splicing, ribosome biogenesis, and RNA degradation. They are superfamily (SF)2 helicases that, similar to SF1, do not form toroidal structures like SF3-6 helicases. Their helicase core consists of two similar protein domains that resemble the fold of the recombination protein RecA. This model describes the C-terminal domain, also called HelicC. |
| 185609 | PTZ00424 | 3.00e-23 | 579 | 722 | 176 | 348 | helicase 45; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.74e-92 | 49 | 282 | 69 | 313 | |
| 1.29e-17 | 56 | 247 | 540 | 756 | |
| 1.29e-17 | 56 | 247 | 540 | 756 | |
| 2.13e-16 | 55 | 267 | 149 | 370 | |
| 2.13e-16 | 55 | 267 | 149 | 370 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.51e-19 | 564 | 719 | 152 | 332 | crystal structure of the eIF4A-PDCD4 complex [Homo sapiens],2ZU6_C crystal structure of the eIF4A-PDCD4 complex [Homo sapiens],2ZU6_D crystal structure of the eIF4A-PDCD4 complex [Homo sapiens],2ZU6_F crystal structure of the eIF4A-PDCD4 complex [Homo sapiens] |
|
| 7.59e-19 | 564 | 719 | 153 | 333 | Chain A, Eukaryotic initiation factor 4A-I [Mus musculus] |
|
| 8.01e-19 | 564 | 719 | 158 | 338 | Chain B, Eukaryotic initiation factor 4A-I [Homo sapiens],7PQ0_B Chain B, Eukaryotic initiation factor 4A-I [Homo sapiens] |
|
| 8.01e-19 | 564 | 719 | 158 | 338 | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex [Homo sapiens] |
|
| 9.05e-19 | 564 | 719 | 170 | 350 | Structure of a human 48S translational initiation complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.87e-48 | 577 | 722 | 266 | 442 | ATP-dependent RNA helicase ROK1 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=ROK1 PE=3 SV=2 |
|
| 2.01e-47 | 577 | 722 | 273 | 449 | ATP-dependent RNA helicase ROK1 OS=Scheffersomyces stipitis (strain ATCC 58785 / CBS 6054 / NBRC 10063 / NRRL Y-11545) OX=322104 GN=ROK1 PE=3 SV=1 |
|
| 4.55e-47 | 577 | 722 | 271 | 447 | ATP-dependent RNA helicase ROK1 OS=Lodderomyces elongisporus (strain ATCC 11503 / CBS 2605 / JCM 1781 / NBRC 1676 / NRRL YB-4239) OX=379508 GN=ROK1 PE=3 SV=1 |
|
| 8.05e-46 | 511 | 721 | 169 | 520 | Probable ATP-dependent RNA helicase ddx52 OS=Dictyostelium discoideum OX=44689 GN=ddx52 PE=3 SV=1 |
|
| 3.37e-45 | 577 | 722 | 283 | 459 | ATP-dependent RNA helicase CHR1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CHR1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
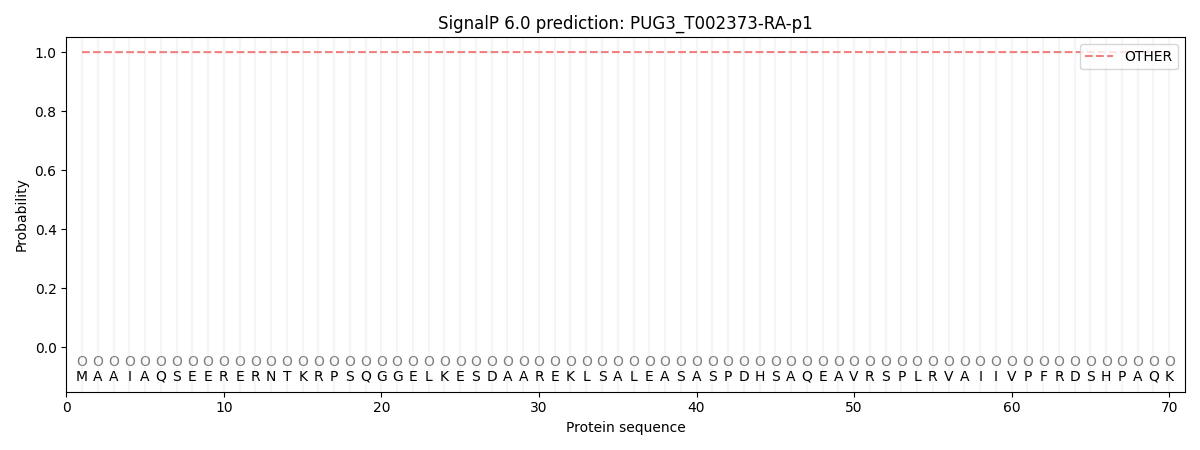
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999889 | 0.000151 |
