You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T002074-RA-p1
You are here: Home > Sequence: PUG3_T002074-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T002074-RA-p1 | |||||||||||
| CAZy Family | AA6 | |||||||||||
| CAZyme Description | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT20 | 2 | 210 | 3.9e-46 | 0.47157894736842104 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 177855 | PLN02205 | 2.58e-109 | 1 | 527 | 307 | 850 | alpha,alpha-trehalose-phosphate synthase [UDP-forming] |
| 184712 | PRK14501 | 4.47e-81 | 1 | 521 | 235 | 723 | putative bifunctional trehalose-6-phosphate synthase/HAD hydrolase subfamily IIB; Provisional |
| 215555 | PLN03063 | 4.48e-69 | 1 | 533 | 249 | 795 | alpha,alpha-trehalose-phosphate synthase (UDP-forming); Provisional |
| 215556 | PLN03064 | 7.41e-60 | 3 | 521 | 335 | 926 | alpha,alpha-trehalose-phosphate synthase (UDP-forming); Provisional |
| 396780 | Trehalose_PPase | 1.14e-53 | 261 | 510 | 1 | 231 | Trehalose-phosphatase. This family consist of trehalose-phosphatases EC:3.1.3.12 these enzyme catalyze the de-phosphorylation of trehalose-6-phosphate to trehalose and orthophosphate. The aligned region is present in trehalose-phosphatases and comprises the entire length of the protein it is also found in the C-terminus of trehalose-6-phosphate synthase EC:2.4.1.15 adjacent to the trehalose-6-phosphate synthase domain - pfam00982. It would appear that the two equivalent genes in the E. coli otsBA operon otsA the trehalose-6-phosphate synthase and otsB trehalose-phosphatase (this family) have undergone gene fusion in most eukaryotes. Trehalose is a common disaccharide of bacteria, fungi and invertebrates that appears to play a major role in desiccation tolerance. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 576 | 291 | 882 | |
| 9.00e-249 | 1 | 538 | 287 | 843 | |
| 1.51e-101 | 1 | 532 | 303 | 849 | |
| 1.51e-101 | 1 | 532 | 303 | 849 | |
| 4.13e-101 | 1 | 542 | 303 | 859 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.69e-38 | 243 | 521 | 6 | 269 | Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 2 [Aspergillus fumigatus Af293],5DXO_A Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 3 [Aspergillus fumigatus Af293],5DXO_B Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 3 [Aspergillus fumigatus Af293] |
|
| 6.12e-37 | 243 | 521 | 6 | 269 | Structure of Aspergillus fumigatus trehalose-6-phosphate phosphatase crystal form 1 [Aspergillus fumigatus Af293] |
|
| 1.50e-31 | 243 | 478 | 6 | 223 | Chain A, trehalose-6-phosphate phosphatase [Cryptococcus neoformans] |
|
| 9.39e-28 | 252 | 481 | 17 | 247 | Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain [Candida albicans SC5314],5DXI_B Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain [Candida albicans SC5314] |
|
| 3.28e-24 | 1 | 219 | 242 | 476 | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose [Candida albicans SC5314],5HUT_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose [Candida albicans SC5314],5HUU_A Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and glucose-6-phosphate [Candida albicans SC5314],5HUU_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and glucose-6-phosphate [Candida albicans SC5314],5HVL_A Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A [Candida albicans SC5314],5HVL_B Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP and validoxylamine A [Candida albicans SC5314] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.10e-99 | 1 | 532 | 304 | 850 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 OS=Arabidopsis thaliana OX=3702 GN=TPS9 PE=2 SV=1 |
|
| 4.04e-97 | 1 | 530 | 304 | 848 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 10 OS=Arabidopsis thaliana OX=3702 GN=TPS10 PE=2 SV=1 |
|
| 5.11e-97 | 7 | 521 | 308 | 834 | Probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 8 OS=Arabidopsis thaliana OX=3702 GN=TPS8 PE=2 SV=1 |
|
| 1.16e-94 | 1 | 525 | 312 | 854 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 OS=Arabidopsis thaliana OX=3702 GN=TPS6 PE=1 SV=2 |
|
| 6.39e-94 | 1 | 526 | 304 | 844 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 5 OS=Arabidopsis thaliana OX=3702 GN=TPS5 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
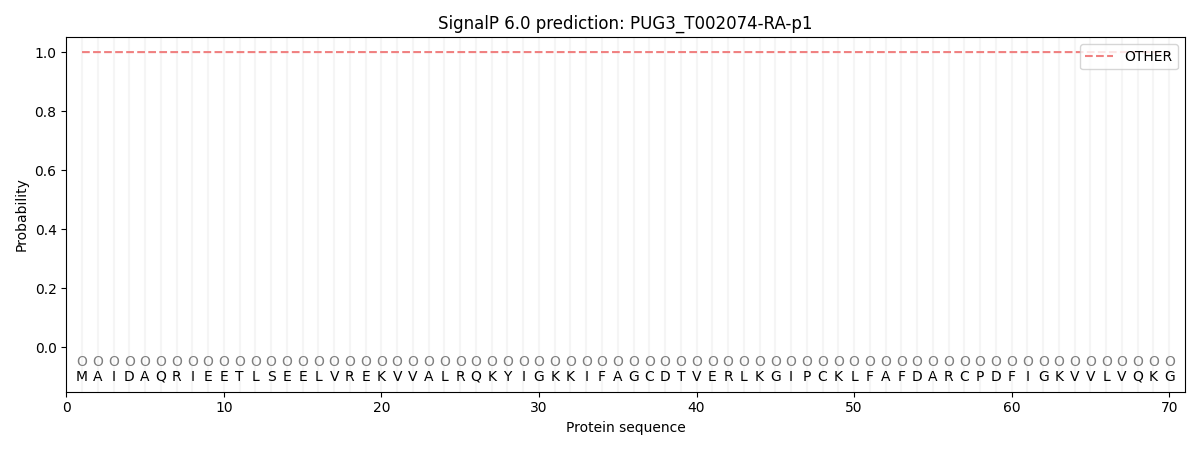
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000052 | 0.000000 |
