You are browsing environment: FUNGIDB
CAZyme Information: PUG3_T001507-RA-p1
You are here: Home > Sequence: PUG3_T001507-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium ultimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium ultimum | |||||||||||
| CAZyme ID | PUG3_T001507-RA-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | Methyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7358; End:10972 Strand: + | |||||||||||
Full Sequence Download help
| YAELLRQTHS STEQQSVNEL KQQAIRHYER ALNADLLWND GVTDNTTNLV LQNRFHVLNT | 60 |
| IGMLLMDLHE LEASISFFER AAAADPTAVH PLGNMALALI NMGEMEAALR INEQALRLDP | 120 |
| NRSHLLHNMG NILHALGRED EALEHFKHAY QLNPNEPETL NSIAIYYAVN GDMNEALQFL | 180 |
| QRTFAIAEAA LAENPSHYEA IIAYNCAQLR LANARLPLVY QSVDEIAEAR SQYMSSLEAL | 240 |
| LNESSAPLVL PDPVNSVSIG SMGYYAVYQG FNDVGIRSQL TRVYRKGAPT LSFTAPHVLH | 300 |
| DRNKTYQFTF PSPPSSRMNR KSRRIAKRYT KYSNETVHVN RRIRVGFHSG LMRQHSVGLL | 360 |
| TQGVITKLPR DAFEVIVIIY DNDERDDLTE LVLRSADSVV FLRQELDQAQ RQISDLELDV | 420 |
| LVFTEIGMHI RTYFLAFSRL ALRTAMFWGH AVTSGIDSVD YFVSSKRFHD DRAEKLFTNK | 480 |
| SQGDVTEQQS KYTECVYEMD HLTTYFFEPL ASQDQLTPPF HALLRESLGL PPKDVLPIMI | 540 |
| LVPQTLYKLH PDFDRLIERV LASVPETSFL AVPIGFMPWL ADVYKRVYFL RRLNTTEFLD | 600 |
| VCAMADLVLD PFPVGGGRSS FEIFAVGTPI VMLAPRTTIL QLTEAMYNVM GVADLIAYSE | 660 |
| DEFVEITVLL AQDEKLRTQA RELILKHKHK LYENAVVIRE WETFLISILA AKPPQERASD | 720 |
| HATSAARVTG ECPHALPLIP VNEPFFELNV FLFGVVDHDN HLVGVTLTLP SEEDDPFEVS | 780 |
| EELAERFEPP LDSLQQNFVA KTMWNAKHRQ SQPVVMTVNV SVTANVYVPV EIRYGDDLHV | 840 |
| ALLMHLEKHR SKMDERIHSV WQIPQVIEVA AGQLQQLLPE YSSPAWIAAR SFPLRFSNAS | 900 |
| NHDAQRNARR ECVTLVMTTC KRLSLFLRTI ASLQRALGIA LESDWSKWFC HLLIVDDNSS | 960 |
| PADRDAMKAH LPQSVEFLYK TPDQRGYAKS LNLAIGRVQS QFVFYLEDDW EFVNYTTREG | 1020 |
| NDTSSASTFL DDALTVLRDN SRREYEPLAQ VLLKNYEGGW SRQLGSIRYF VHEFAASDPT | 1080 |
| LSFAFWPGFS LNPGLWDLGA ILEAMEWDDT IDRPRQRPIF DEVSDRFETL FACELWRTGL | 1140 |
| RVAYLDGEFI SHTGAPLGTN ASAYVLNDLP RRFDFVESFA RSAFPLR | 1187 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 113 | 707 | 1.1e-62 | 0.5617021276595745 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 2.30e-32 | 104 | 684 | 6 | 588 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 5.79e-19 | 92 | 182 | 4 | 94 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 2.11e-18 | 57 | 151 | 3 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 4.14e-13 | 125 | 238 | 3 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 223533 | TPR | 4.43e-13 | 64 | 204 | 139 | 276 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ32129.1|GT41 | 1.15e-54 | 208 | 724 | 480 | 938 |
| QEG14440.1|GT41 | 1.15e-54 | 208 | 724 | 480 | 938 |
| QDU12502.1|GT41 | 1.15e-54 | 208 | 724 | 480 | 938 |
| BAP54338.1|GT41 | 3.68e-54 | 208 | 707 | 138 | 584 |
| QDT18528.1|GT41 | 9.76e-53 | 216 | 727 | 488 | 935 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5DJS_A | 6.14e-10 | 340 | 682 | 160 | 487 | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_B Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_C Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_D Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum] |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
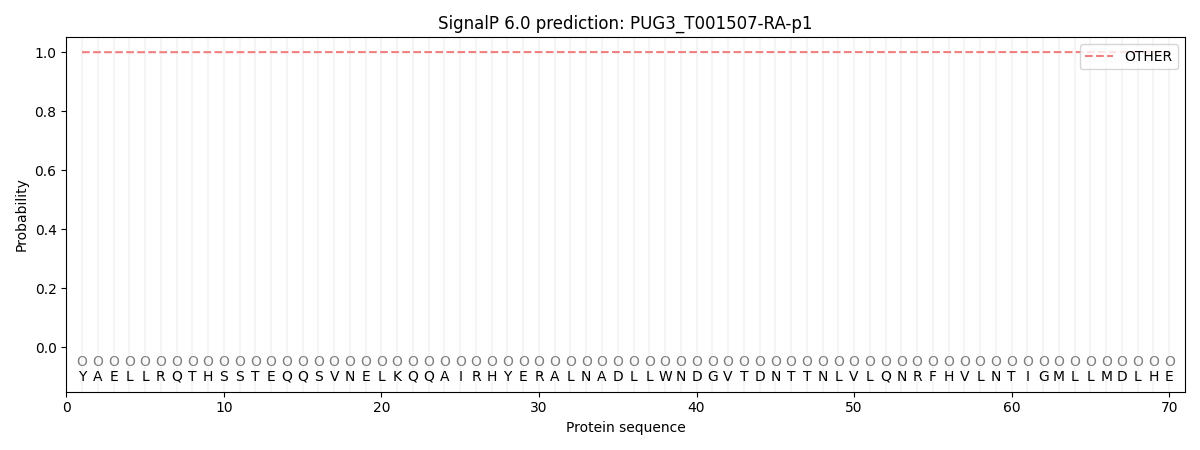
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999778 | 0.000256 |
