You are browsing environment: FUNGIDB
CAZyme Information: PTTG_29564-t43_1-p1
You are here: Home > Sequence: PTTG_29564-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
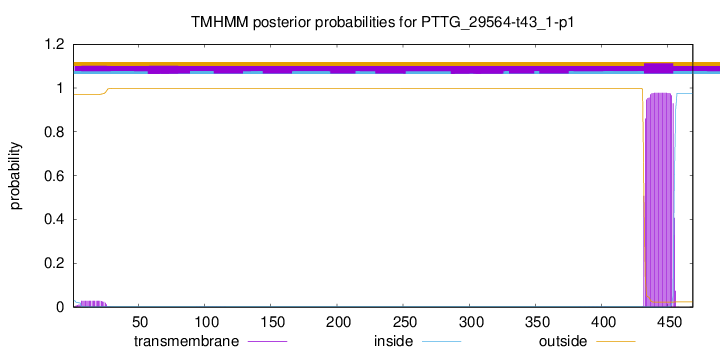
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_29564-t43_1-p1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226609 | ManB2 | 4.16e-05 | 128 | 248 | 177 | 272 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.05e-153 | 160 | 465 | 1 | 306 | |
| 8.49e-131 | 10 | 420 | 25 | 439 | |
| 1.24e-111 | 12 | 385 | 16 | 394 | |
| 3.76e-44 | 91 | 363 | 68 | 342 | |
| 6.47e-44 | 92 | 363 | 123 | 396 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.69e-07 | 92 | 246 | 70 | 201 | Crystal structure of beta-1,3-xylanase from Vibrio sp. AX-4 [Vibrio sp. AX-4],3VPL_A Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase [Vibrio sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.63e-06 | 92 | 246 | 92 | 223 | Beta-1,3-xylanase XYL4 OS=Vibrio sp. OX=678 GN=xyl4 PE=1 SV=1 |
|
| 5.34e-06 | 86 | 246 | 86 | 223 | Beta-1,3-xylanase TXYA OS=Vibrio sp. OX=678 GN=txyA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
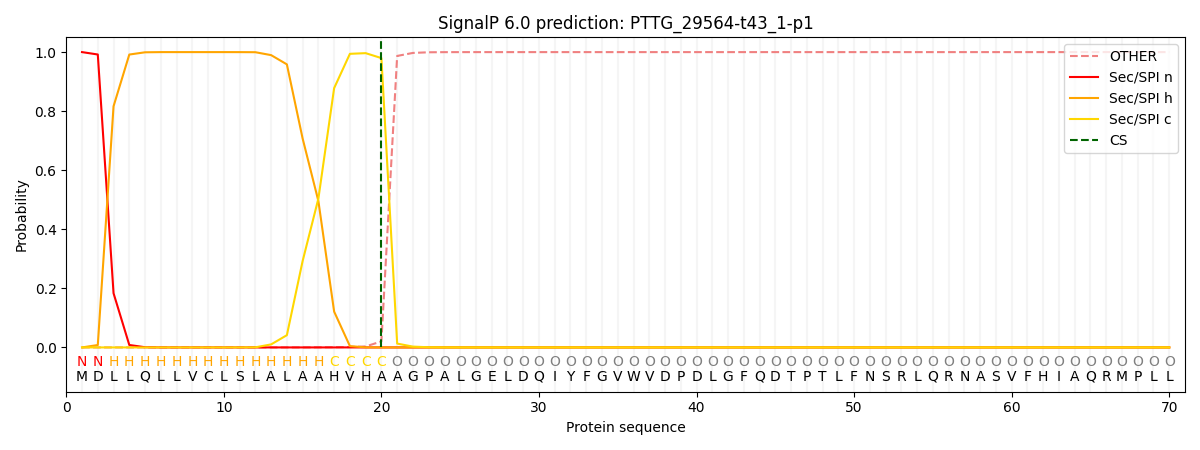
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000185 | 0.999793 | CS pos: 20-21. Pr: 0.9797 |