You are browsing environment: FUNGIDB
CAZyme Information: PTTG_09892-t43_1-p1
You are here: Home > Sequence: PTTG_09892-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
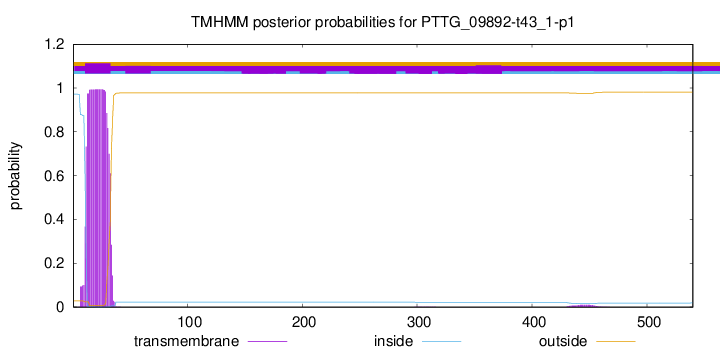
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_09892-t43_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT49 | 212 | 496 | 3.3e-42 | 0.8338278931750742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404735 | Glyco_transf_49 | 1.65e-55 | 217 | 494 | 1 | 326 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.10e-166 | 6 | 527 | 18 | 548 | |
| 2.04e-132 | 176 | 538 | 79 | 441 | |
| 8.77e-132 | 180 | 524 | 53 | 389 | |
| 1.03e-131 | 133 | 528 | 45 | 427 | |
| 4.24e-130 | 179 | 528 | 83 | 424 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.13e-22 | 207 | 466 | 392 | 639 | Xylosyl- and glucuronyltransferase LARGE2 OS=Mus musculus OX=10090 GN=Large2 PE=1 SV=1 |
|
| 3.13e-22 | 215 | 466 | 399 | 639 | Xylosyl- and glucuronyltransferase LARGE2 OS=Rattus norvegicus OX=10116 GN=Large2 PE=2 SV=1 |
|
| 8.72e-21 | 218 | 465 | 348 | 584 | Glycosyltransferase-like protein gnt14 OS=Dictyostelium discoideum OX=44689 GN=gnt14 PE=2 SV=1 |
|
| 4.35e-20 | 214 | 466 | 470 | 709 | Xylosyl- and glucuronyltransferase LARGE1 OS=Mus musculus OX=10090 GN=Large1 PE=1 SV=1 |
|
| 6.28e-20 | 215 | 465 | 350 | 592 | Glycosyltransferase-like protein LARGE OS=Caenorhabditis elegans OX=6239 GN=lge-1 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000012 | 0.000001 |