You are browsing environment: FUNGIDB
CAZyme Information: PTTG_09357-t43_1-p1
You are here: Home > Sequence: PTTG_09357-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_09357-t43_1-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 131 | 279 | 7.8e-52 | 0.7700534759358288 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 2.09e-35 | 138 | 341 | 10 | 188 | Amb_all domain. |
| 226384 | PelB | 1.06e-15 | 119 | 341 | 84 | 275 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 3.81e-05 | 133 | 237 | 23 | 136 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 404168 | Beta_helix | 8.78e-04 | 157 | 268 | 21 | 110 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| 404168 | Beta_helix | 0.008 | 161 | 260 | 2 | 79 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.46e-70 | 41 | 428 | 30 | 385 | |
| 1.07e-67 | 36 | 381 | 23 | 331 | |
| 4.03e-67 | 43 | 381 | 27 | 330 | |
| 1.43e-66 | 42 | 428 | 25 | 380 | |
| 2.58e-66 | 41 | 427 | 23 | 376 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-61 | 43 | 381 | 9 | 314 | Pectin Lyase B [Aspergillus niger] |
|
| 1.15e-55 | 43 | 422 | 9 | 359 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 1.15e-55 | 43 | 422 | 9 | 359 | Pectin Lyase A [Aspergillus niger] |
|
| 4.22e-07 | 147 | 289 | 82 | 212 | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
|
| 4.29e-07 | 44 | 240 | 17 | 253 | Structure of the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.97e-68 | 5 | 420 | 4 | 376 | Probable pectin lyase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelA PE=3 SV=1 |
|
| 5.56e-68 | 43 | 420 | 27 | 376 | Probable pectin lyase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=pelA PE=3 SV=1 |
|
| 4.71e-65 | 43 | 422 | 27 | 378 | Probable pectin lyase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pelB PE=3 SV=1 |
|
| 4.71e-65 | 43 | 422 | 27 | 378 | Probable pectin lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelA PE=3 SV=1 |
|
| 9.13e-65 | 40 | 421 | 26 | 386 | Probable pectin lyase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pelF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
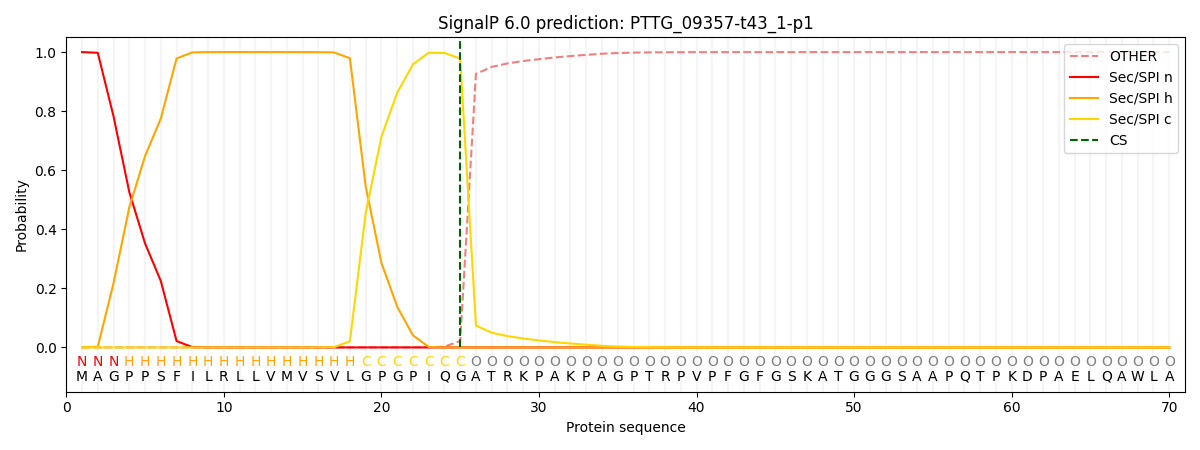
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000302 | 0.999666 | CS pos: 25-26. Pr: 0.9781 |
