You are browsing environment: FUNGIDB
CAZyme Information: PTTG_09307-t43_1-p1
You are here: Home > Sequence: PTTG_09307-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_09307-t43_1-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:15 | 3.2.1.151:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 145 | 301 | 1.2e-28 | 0.9743589743589743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 5.44e-07 | 72 | 291 | 2 | 194 | Glycosyl hydrolase family 12. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.18e-23 | 72 | 302 | 52 | 260 | |
| 2.51e-19 | 75 | 300 | 49 | 248 | |
| 6.76e-19 | 71 | 302 | 46 | 251 | |
| 3.07e-18 | 125 | 302 | 68 | 244 | |
| 3.07e-18 | 125 | 302 | 68 | 244 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.19e-16 | 75 | 302 | 41 | 242 | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus],4NPR_B Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus] |
|
| 1.93e-06 | 75 | 266 | 19 | 184 | The Humicola grisea Cel12A Enzyme Structure at 1.2 A Resolution [Trichocladium griseum],1UU4_A X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH CELLOBIOSE [Trichocladium griseum],1UU5_A X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE [Trichocladium griseum],1UU6_A X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED CELLOPENTAOSE [Trichocladium griseum],1W2U_A X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED THIO CELLOTETRAOSE [Trichocladium griseum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.62e-18 | 75 | 302 | 36 | 240 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=xgeA PE=3 SV=1 |
|
| 2.28e-17 | 75 | 302 | 36 | 237 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xgeA PE=3 SV=1 |
|
| 4.23e-16 | 71 | 302 | 34 | 239 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xgeA PE=3 SV=1 |
|
| 4.23e-16 | 71 | 302 | 34 | 239 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xgeA PE=3 SV=1 |
|
| 5.63e-16 | 75 | 302 | 36 | 237 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xgeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
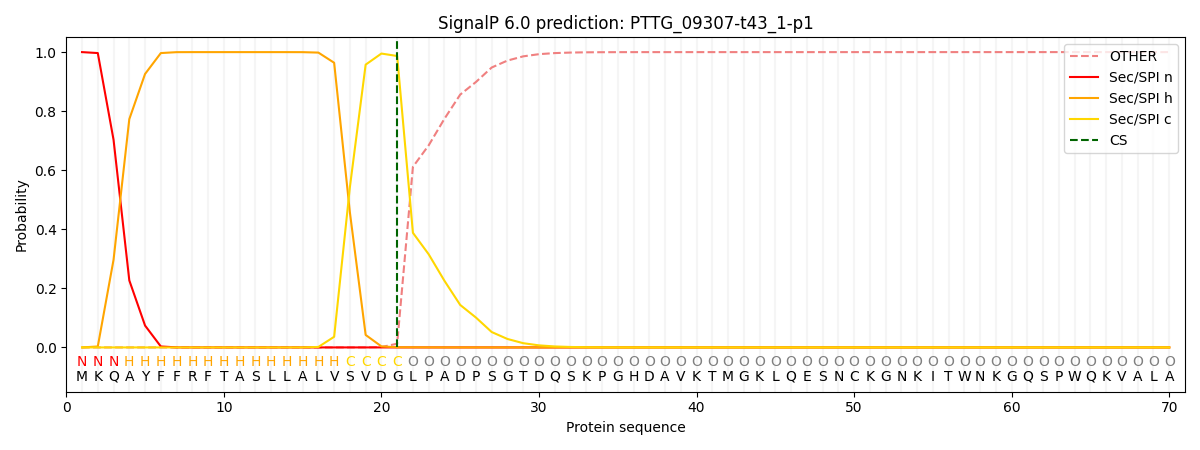
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000238 | 0.999722 | CS pos: 21-22. Pr: 0.9868 |
