You are browsing environment: FUNGIDB
CAZyme Information: PTTG_08591-t43_1-p1
Basic Information
help
| Species |
Puccinia triticina
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina
|
| CAZyme ID |
PTTG_08591-t43_1-p1
|
| CAZy Family |
GT90 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Ptriticina1-1BBBDRace1 |
15692 |
630390 |
814 |
14878
|
|
| Gene Location |
No EC number prediction in PTTG_08591-t43_1-p1.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 236776
|
PRK10856 |
0.002 |
61 |
147 |
168 |
254 |
cytoskeleton protein RodZ. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.89e-133 |
142 |
423 |
180 |
458 |
| 8.91e-67 |
143 |
396 |
16 |
279 |
| 4.41e-66 |
151 |
396 |
21 |
278 |
| 8.97e-66 |
151 |
396 |
21 |
278 |
| 5.25e-64 |
143 |
396 |
13 |
276 |
PTTG_08591-t43_1-p1 has no PDB hit.
PTTG_08591-t43_1-p1 has no Swissprot hit.
This protein is predicted as SP
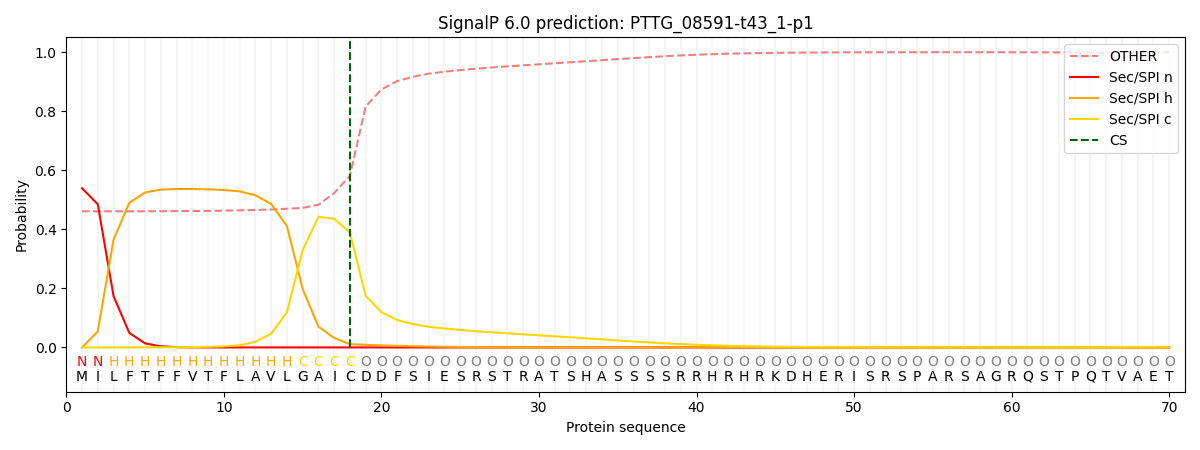
| Other |
SP_Sec_SPI |
CS Position |
| 0.477214 |
0.522770 |
CS pos: 18-19. Pr: 0.3895 |
There is no transmembrane helices in PTTG_08591-t43_1-p1.

