You are browsing environment: FUNGIDB
CAZyme Information: PTTG_06227-t43_1-p1
You are here: Home > Sequence: PTTG_06227-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_06227-t43_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.25:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 70 | 858 | 1.1e-66 | 0.6409574468085106 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 1.72e-40 | 94 | 794 | 65 | 628 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 407658 | Mannosidase_ig | 5.18e-16 | 809 | 902 | 1 | 91 | Mannosidase Ig/CBM-like domain. This domain corresponds to domain 4 in the structure of Bacteroides thetaiotaomicron beta-mannosidase, BtMan2A. This domain has an Ig-like fold. |
| 407625 | Ig_mannosidase | 1.40e-10 | 917 | 996 | 12 | 78 | Ig-fold domain. This Ig-like fold domain is found in mannosidase enzymes. |
| 397119 | Glyco_hydro_2_C | 2.39e-04 | 438 | 620 | 8 | 185 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| 397120 | Glyco_hydro_2_N | 0.009 | 40 | 184 | 21 | 154 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.87e-192 | 37 | 998 | 23 | 922 | |
| 1.46e-190 | 21 | 998 | 5 | 929 | |
| 1.46e-190 | 21 | 998 | 5 | 929 | |
| 2.61e-190 | 36 | 998 | 23 | 925 | |
| 2.61e-190 | 36 | 998 | 23 | 925 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.02e-175 | 12 | 996 | 7 | 938 | Structure of Fungal beta-mannosidase from Glycoside Hydrolase Family 2 of Trichoderma harzianum [Trichoderma harzianum],4UOJ_A Chain A, BETA-MANNOSIDASE GH2 [Trichoderma harzianum],4UOJ_B Chain B, BETA-MANNOSIDASE GH2 [Trichoderma harzianum] |
|
| 3.24e-91 | 41 | 819 | 15 | 699 | mouse beta-mannosidase (MANBA) [Mus musculus],6DDU_A mouse beta-mannosidase bound to beta-D-mannose (MANBA) [Mus musculus] |
|
| 1.76e-68 | 44 | 910 | 15 | 764 | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VJX_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VL4_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VMF_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VO5_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VOT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron VPI-5482],2VQT_A Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VQT_B Structural and biochemical evidence for a boat-like transition state in beta-mannosidases [Bacteroides thetaiotaomicron],2VR4_A Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482],2VR4_B Transition-state mimicry in mannoside hydrolysis: characterisation of twenty six inhibitors and insight into binding from linear free energy relationships and 3-D structure [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.81e-68 | 44 | 910 | 17 | 766 | Structure of a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JE8_B Structure of a beta-mannosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.83e-68 | 44 | 910 | 17 | 766 | Chain A, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP6_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_A Chain A, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482],7OP7_B Chain B, Beta-mannosidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.85e-191 | 20 | 996 | 8 | 927 | Beta-mannosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=mndA PE=3 SV=2 |
|
| 4.63e-191 | 36 | 998 | 23 | 925 | Beta-mannosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mndA PE=3 SV=1 |
|
| 8.52e-189 | 20 | 998 | 4 | 928 | Beta-mannosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=mndA PE=3 SV=1 |
|
| 9.10e-183 | 20 | 998 | 7 | 923 | Beta-mannosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndA PE=3 SV=1 |
|
| 3.57e-182 | 20 | 998 | 7 | 923 | Beta-mannosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=mndA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
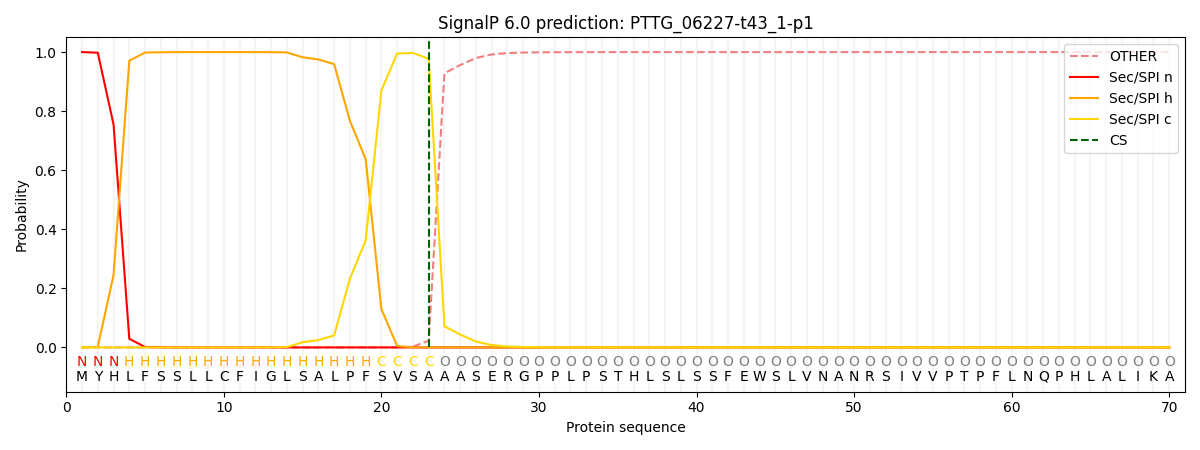
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000283 | 0.999695 | CS pos: 23-24. Pr: 0.9773 |
