You are browsing environment: FUNGIDB
CAZyme Information: PTTG_05925-t43_1-p1
You are here: Home > Sequence: PTTG_05925-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia triticina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina | |||||||||||
| CAZyme ID | PTTG_05925-t43_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 974 | 1183 | 1.9e-27 | 0.9898477157360406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404513 | Glyco_trans_2_3 | 4.69e-26 | 974 | 1184 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 376607 | Acyl_transf_3 | 9.78e-09 | 295 | 692 | 1 | 330 | Acyltransferase family. This family includes a range of acyltransferase enzymes. This domain is found in many as yet uncharacterized C. elegans proteins and it is approximately 300 amino acids long. |
| 224748 | OafA | 3.51e-08 | 290 | 696 | 10 | 355 | Peptidoglycan/LPS O-acetylase OafA/YrhL, contains acyltransferase and SGNH-hydrolase domains [Cell wall/membrane/envelope biogenesis]. |
| 404520 | Glyco_tranf_2_3 | 8.13e-07 | 972 | 1117 | 88 | 230 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| 224136 | BcsA | 8.21e-07 | 901 | 1187 | 64 | 348 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 84 | 1236 | 85 | 1230 | |
| 2.84e-222 | 286 | 1236 | 121 | 1070 | |
| 3.16e-216 | 286 | 1236 | 118 | 1067 | |
| 6.43e-216 | 286 | 1236 | 119 | 1068 | |
| 3.55e-215 | 286 | 1236 | 119 | 1068 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.38e-37 | 849 | 1235 | 56 | 449 | Beta-1,4-mannosyltransferase egh OS=Drosophila melanogaster OX=7227 GN=egh PE=2 SV=1 |
|
| 3.98e-34 | 849 | 1118 | 56 | 330 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis briggsae OX=6238 GN=bre-3 PE=3 SV=1 |
|
| 2.37e-33 | 849 | 1118 | 56 | 330 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis elegans OX=6239 GN=bre-3 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000012 |
TMHMM Annotations download full data without filtering help
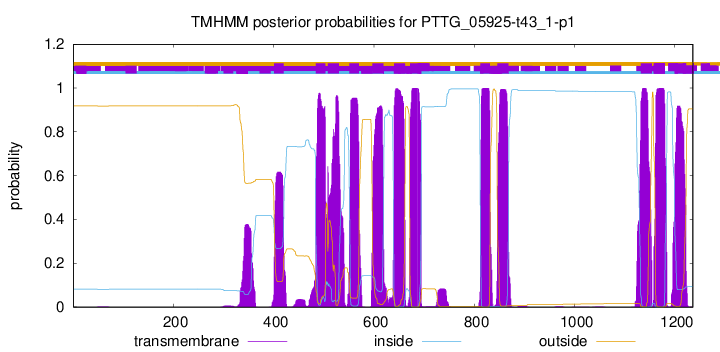
| Start | End |
|---|---|
| 401 | 420 |
| 484 | 503 |
| 508 | 530 |
| 551 | 570 |
| 597 | 619 |
| 640 | 662 |
| 672 | 694 |
| 812 | 831 |
| 846 | 868 |
| 1132 | 1154 |
| 1158 | 1180 |
| 1200 | 1222 |
