You are browsing environment: FUNGIDB
CAZyme Information: PTTG_01499-t43_1-p1
Basic Information
help
| Species |
Puccinia triticina
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia triticina
|
| CAZyme ID |
PTTG_01499-t43_1-p1
|
| CAZy Family |
CE5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 822 |
|
92468.44 |
7.1584 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Ptriticina1-1BBBDRace1 |
15692 |
630390 |
814 |
14878
|
|
| Gene Location |
No EC number prediction in PTTG_01499-t43_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT90 |
421 |
799 |
1.5e-50 |
0.924 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 214773
|
CAP10 |
9.35e-11 |
426 |
675 |
28 |
220 |
Putative lipopolysaccharide-modifying enzyme. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.46e-178 |
122 |
805 |
159 |
750 |
| 2.45e-81 |
124 |
807 |
67 |
637 |
| 2.45e-81 |
124 |
807 |
67 |
637 |
| 2.89e-81 |
124 |
807 |
64 |
630 |
| 3.40e-81 |
124 |
807 |
67 |
637 |
PTTG_01499-t43_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.22e-24 |
124 |
802 |
152 |
659 |
Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
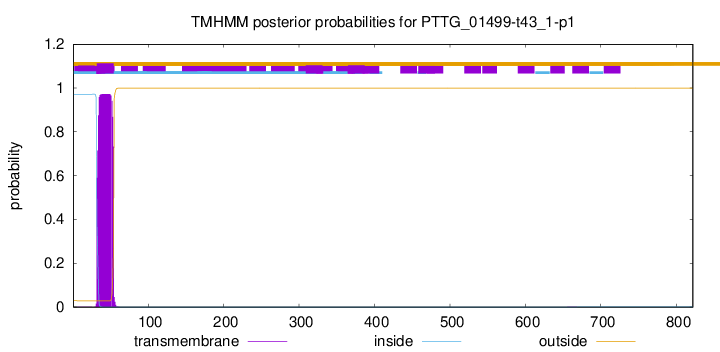
This protein is predicted as OTHER
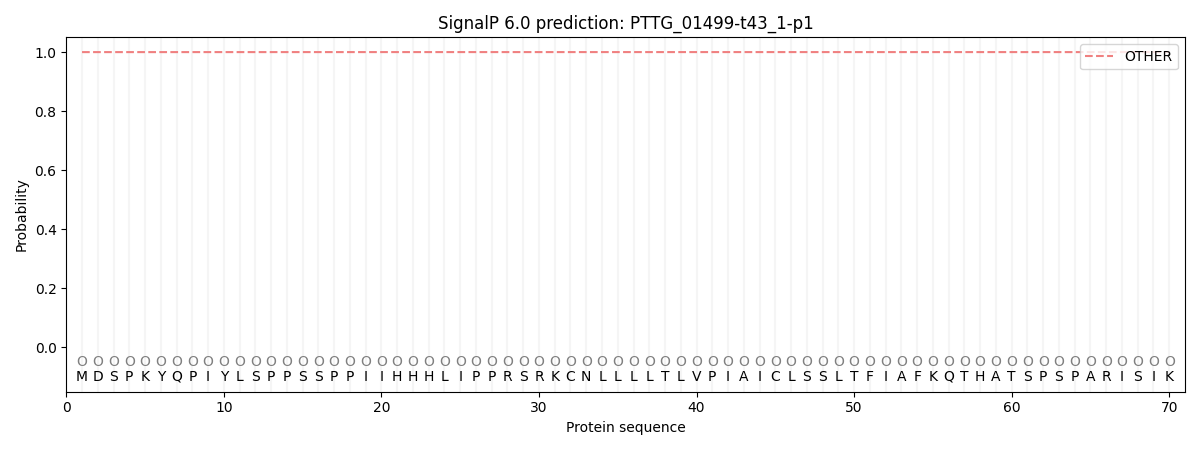
| Other |
SP_Sec_SPI |
CS Position |
| 1.000025 |
0.000019 |
|