You are browsing environment: FUNGIDB
CAZyme Information: PSURA_94521T0-p1
You are here: Home > Sequence: PSURA_94521T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_94521T0-p1 | |||||||||||
| CAZy Family | PL3 | |||||||||||
| CAZyme Description | Translation initiation factor 2C (eIF-2C) and related proteins | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 31 | 500 | 2.3e-133 | 0.9813519813519813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 240015 | Piwi_ago-like | 1.02e-169 | 1394 | 1818 | 1 | 426 | Piwi_ago-like: PIWI domain, Argonaute-like subfamily. Argonaute is the central component of the RNA-induced silencing complex (RISC) and related complexes. The PIWI domain is the C-terminal portion of Argonaute and consists of two subdomains, one of which provides the 5' anchoring of the guide RNA and the other, the catalytic site for slicing. |
| 395176 | Glyco_hydro_1 | 2.76e-135 | 32 | 502 | 5 | 451 | Glycosyl hydrolase family 1. |
| 215631 | PLN03202 | 2.15e-127 | 965 | 1853 | 36 | 899 | protein argonaute; Provisional |
| 225343 | BglB | 1.88e-108 | 32 | 505 | 4 | 456 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 396649 | Piwi | 6.95e-101 | 1520 | 1819 | 1 | 296 | Piwi domain. This domain is found in the protein Piwi and its relatives. The function of this domain is the dsRNA guided hydrolysis of ssRNA. Determination of the crystal structure of Argonaute reveals that PIWI is an RNase H domain, and identifies Argonaute as Slicer, the enzyme that cleaves mRNA in the RNAi RISC complex. In addition, Mg+2 dependence and production of 3'-OH and 5' phosphate products are shared characteristics of RNaseH and RISC. The PIWI domain core has a tertiary structure belonging to the RNase H family of enzymes. RNase H fold proteins all have a five-stranded mixed beta-sheet surrounded by helices. By analogy to RNase H enzymes which cleave single-stranded RNA guided by the DNA strand in an RNA/DNA hybrid, the PIWI domain can be inferred to cleave single-stranded RNA, for example mRNA, guided by double stranded siRNA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.28e-205 | 23 | 504 | 23 | 515 | |
| 7.74e-204 | 28 | 504 | 9 | 496 | |
| 1.00e-203 | 81 | 537 | 6 | 451 | |
| 9.87e-191 | 30 | 505 | 21 | 507 | |
| 1.30e-95 | 30 | 495 | 2 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.28e-141 | 1052 | 1853 | 62 | 860 | Human Argonaute-2 - miR-20a complex [Homo sapiens] |
|
| 1.66e-141 | 1052 | 1853 | 60 | 858 | Human Argonaute2 A481T Mutant Bound to t1-A Target RNA [Homo sapiens],4Z4I_A Human Argonaute2 A481T Mutant Bound to t1-G Target RNA [Homo sapiens] |
|
| 1.66e-141 | 1052 | 1853 | 60 | 858 | Crystal Structure of Human Argonaute2 [Homo sapiens],4OLB_A Crystal Structure of Human Argonaute2 Bound to Tryptophan [Homo sapiens],4W5N_A The Crystal Structure of Human Argonaute2 Bound to a Defined Guide RNA [Homo sapiens],4W5O_A The Crystal Structure of Human Argonaute2 Bound to a Guide and Target RNA Containing Seed Pairing from 2-9 [Homo sapiens],4W5Q_A The Crystal Structure of Human Argonaute2 Bound to a Guide and Target RNA Containing Seed Pairing from 2-8 [Homo sapiens],4W5R_A The Crystal Structure of Human Argonaute2 Bound to a Guide and Target RNA Containing Seed Pairing from 2-8 (Long Target) [Homo sapiens],4W5T_A The Crystal Structure of Human Argonaute2 Bound to a Guide and Target RNA Containing Seed Pairing from 2-7 [Homo sapiens],4Z4C_A Human Argonaute2 Bound to t1-C Target RNA [Homo sapiens],4Z4D_A Human Argonaute2 Bound to t1-G Target RNA [Homo sapiens],4Z4E_A Human Argonaute2 Bound to t1-U Target RNA [Homo sapiens],4Z4F_A Human Argonaute2 Bound to t1-DAP Target RNA [Homo sapiens],4Z4G_A Human Argonaute2 Bound to t1-Inosine Target RNA [Homo sapiens],5JS1_A Human Argonaute2 Bound to an siRNA [Homo sapiens],5JS2_A Human Argonaute-2 Bound to a Modified siRNA [Homo sapiens],5KI6_A Human Argonaute-2 bound to a guide RNA with a nucleobase modification at position 1 [Homo sapiens],6CBD_A Crystal Structure of Human Argonaute2 Bound to Three Tryptophans [Homo sapiens] |
|
| 3.42e-141 | 1060 | 1853 | 61 | 861 | Human Argonaute3 bound to guide RNA [Homo sapiens],5VM9_C Human Argonaute3 bound to guide RNA [Homo sapiens] |
|
| 5.98e-141 | 1052 | 1853 | 60 | 858 | Chain A, Protein argonaute-2 [Homo sapiens],7KI3_D Chain D, Protein argonaute-2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.77e-143 | 1061 | 1818 | 167 | 940 | Protein argonaute PNH1 OS=Oryza sativa subsp. japonica OX=39947 GN=PHN1 PE=2 SV=1 |
|
| 6.21e-141 | 1052 | 1853 | 60 | 858 | Protein argonaute-2 OS=Homo sapiens OX=9606 GN=AGO2 PE=1 SV=3 |
|
| 6.39e-141 | 1052 | 1853 | 61 | 859 | Protein argonaute-2 OS=Mus musculus OX=10090 GN=Ago2 PE=1 SV=3 |
|
| 8.79e-141 | 1060 | 1853 | 59 | 859 | Protein argonaute-3 OS=Mus musculus OX=10090 GN=Ago3 PE=2 SV=2 |
|
| 1.67e-140 | 1060 | 1853 | 59 | 859 | Protein argonaute-3 OS=Homo sapiens OX=9606 GN=AGO3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
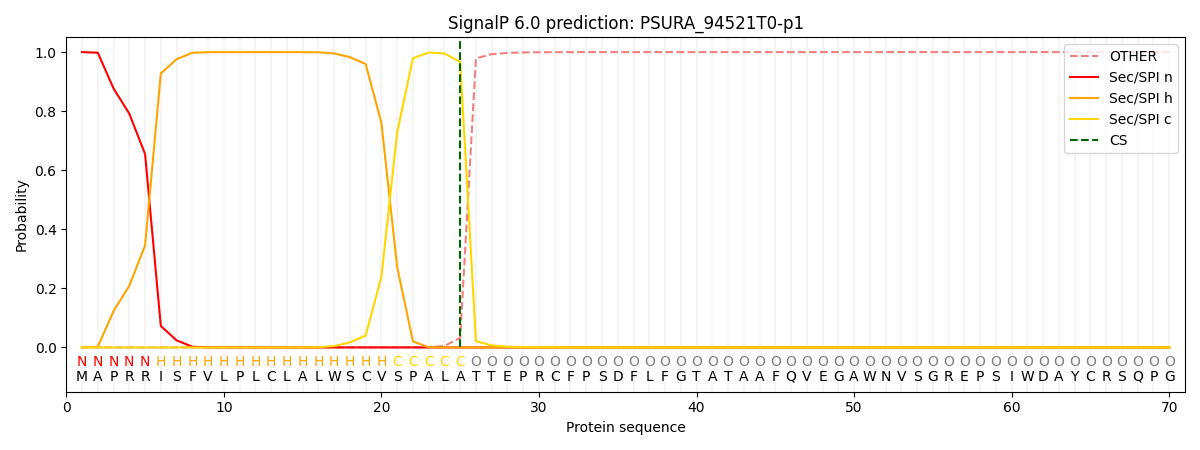
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000321 | 0.999640 | CS pos: 25-26. Pr: 0.9670 |
