You are browsing environment: FUNGIDB
CAZyme Information: PSURA_86725T0-p1
You are here: Home > Sequence: PSURA_86725T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_86725T0-p1 | |||||||||||
| CAZy Family | GT71|GT71|GT71 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.122:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 226 | 422 | 4.5e-36 | 0.9791666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 4.96e-24 | 226 | 424 | 3 | 196 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 215596 | PLN03133 | 4.27e-07 | 204 | 413 | 380 | 579 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-18 | 207 | 428 | 53 | 259 | |
| 1.58e-18 | 209 | 462 | 78 | 324 | |
| 1.58e-18 | 209 | 462 | 78 | 324 | |
| 1.58e-18 | 209 | 462 | 78 | 324 | |
| 1.58e-18 | 209 | 462 | 78 | 324 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.81e-19 | 209 | 462 | 78 | 324 | Beta-1,3-galactosyltransferase 1 OS=Homo sapiens OX=9606 GN=B3GALT1 PE=1 SV=1 |
|
| 2.81e-19 | 209 | 462 | 78 | 324 | Beta-1,3-galactosyltransferase 1 OS=Gorilla gorilla gorilla OX=9595 GN=B3GALT1 PE=3 SV=1 |
|
| 2.81e-19 | 209 | 462 | 78 | 324 | Beta-1,3-galactosyltransferase 1 OS=Pan paniscus OX=9597 GN=B3GALT1 PE=3 SV=1 |
|
| 2.81e-19 | 209 | 462 | 78 | 324 | Beta-1,3-galactosyltransferase 1 OS=Pan troglodytes OX=9598 GN=B3GALT1 PE=3 SV=1 |
|
| 2.81e-19 | 209 | 462 | 78 | 324 | Beta-1,3-galactosyltransferase 1 OS=Mus musculus OX=10090 GN=B3galt1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
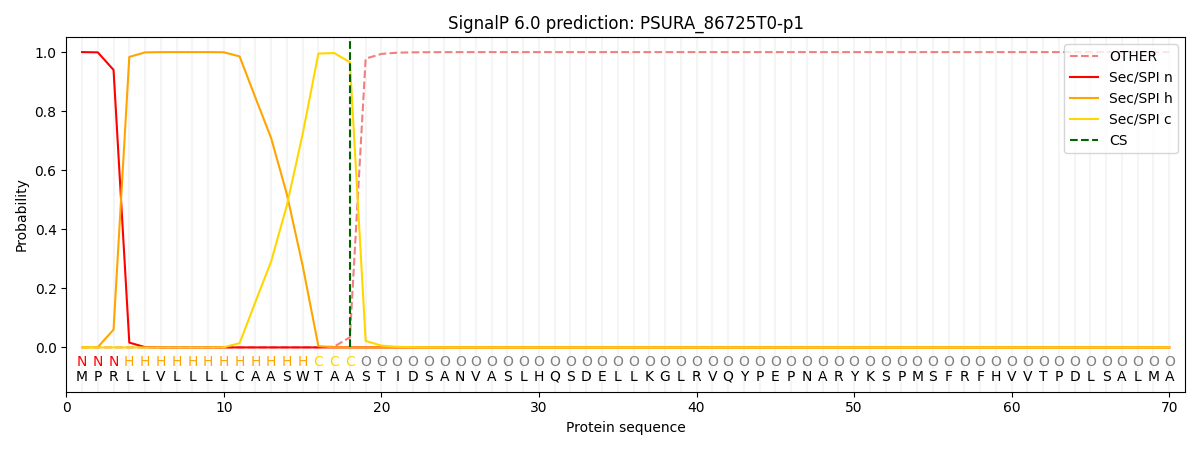
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000238 | 0.999756 | CS pos: 18-19. Pr: 0.9657 |
