You are browsing environment: FUNGIDB
CAZyme Information: PSURA_86595T0-p1
You are here: Home > Sequence: PSURA_86595T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
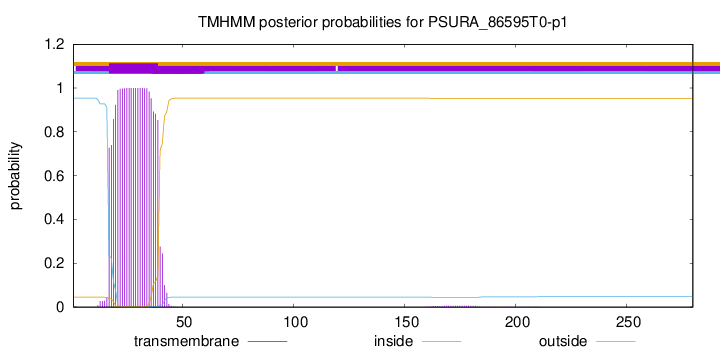
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_86595T0-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:73 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 89 | 256 | 7.6e-41 | 0.3923766816143498 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 240427 | PTZ00470 | 5.10e-50 | 71 | 280 | 61 | 310 | glycoside hydrolase family 47 protein; Provisional |
| 396217 | Glyco_hydro_47 | 3.34e-48 | 91 | 280 | 3 | 236 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.69e-46 | 81 | 279 | 143 | 396 | |
| 1.88e-38 | 19 | 275 | 11 | 292 | |
| 5.16e-33 | 81 | 280 | 110 | 352 | |
| 5.32e-33 | 81 | 280 | 125 | 367 | |
| 2.91e-32 | 47 | 274 | 153 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.77e-32 | 41 | 280 | 60 | 324 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
|
| 8.79e-32 | 81 | 275 | 21 | 257 | Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus],5KKB_B Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus] |
|
| 9.68e-32 | 81 | 275 | 19 | 255 | Structure of mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) [Mus musculus] |
|
| 1.39e-31 | 78 | 280 | 1 | 241 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 1.43e-31 | 78 | 280 | 1 | 241 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.33e-32 | 83 | 279 | 181 | 420 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Mus musculus OX=10090 GN=Man1a2 PE=1 SV=1 |
|
| 1.33e-32 | 81 | 280 | 179 | 421 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB OS=Homo sapiens OX=9606 GN=MAN1A2 PE=1 SV=1 |
|
| 1.90e-31 | 41 | 280 | 221 | 485 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Homo sapiens OX=9606 GN=MAN1B1 PE=1 SV=2 |
|
| 1.49e-30 | 81 | 275 | 196 | 432 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA OS=Mus musculus OX=10090 GN=Man1a1 PE=1 SV=1 |
|
| 1.50e-30 | 80 | 280 | 206 | 444 | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase OS=Mus musculus OX=10090 GN=Man1b1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
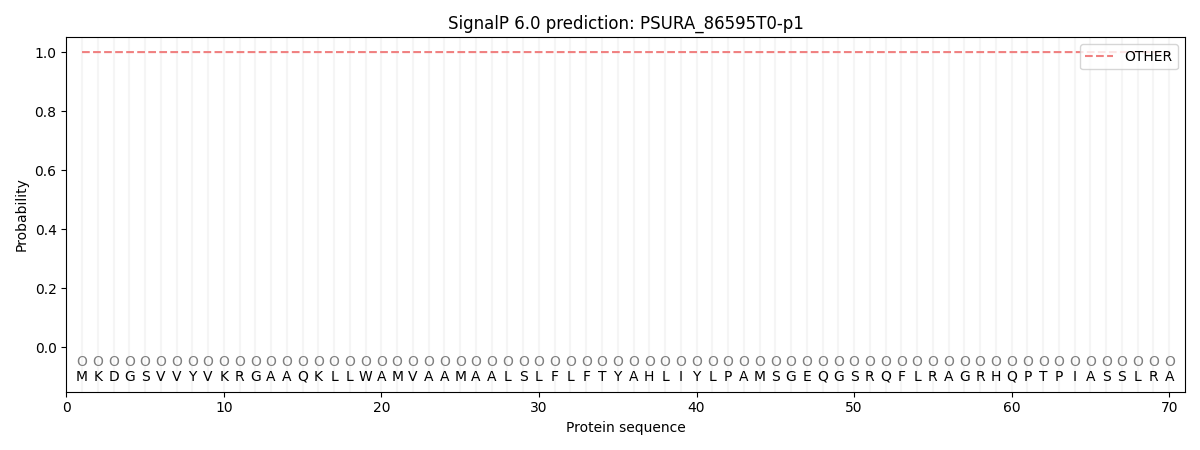
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000050 | 0.000002 |