You are browsing environment: FUNGIDB
CAZyme Information: PSURA_85409T0-p1
You are here: Home > Sequence: PSURA_85409T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_85409T0-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | N-arginine dibasic convertase NRD1 and related Zn2+-dependent endopeptidases, insulinase superfamily | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34968; End:39277 Strand: + | |||||||||||
Full Sequence Download help
| MMKAAIALML GASSLSVATA DPRVMIESTY TDTRGLNDLA QAAQGKYMGT ATDIKQLSDQ | 60 |
| YYTQELNNTK DFSMITPANA MKWDATEAKQ GVFTFDDADK IVAFANATGA RVRCHALVWH | 120 |
| QQVPAWVESL EKAELLEAMS NHITKVMTHF GDSCYSWDVV NEAMDEDGSY RQSFWFEKTG | 180 |
| KEYISAAYKT ANAVKKKLGL KVKLYYNDYN INIANKKSDA VLEMVGALRN VSNWVDGVGF | 240 |
| QSHYSNNDTA STVGSDIFWN LRRFTLSRVD VALTELDVKT SSANPTVTEQ QQQVGIYTNA | 300 |
| VSACKKTKRC VGVTVWDFVD TYSWVEASAP LLYYQPEGAN TPLVRKATYD AVTAGWIFRV | 360 |
| VKLVKFDGME RVVQCDGIDV SSLDERDFKY VTLPNGLHAL VVSDATTETA SAAMDVRVGF | 420 |
| HSDPDAVPGL AHFCEHMLFL GTRKFPDENS YSAFLAANGG SSNAFTAGRD TNFYFDVGAA | 480 |
| HLHEALDRFA QFFIAPLFTP SATEREVNAV DSESTNYLQD DSWRINQLER GLGNVKHPYH | 540 |
| KFGVGNKETL SVTPAEQGID VREQLLTFYH EFYSASIMKL VIYGKEDVET LSQWACELFT | 600 |
| EIPNSMRRSP AFDQEQPYTA DQLARRLEVV PVMDWKVVQV SWVLPPLRGK GYSQQHASVL | 660 |
| SHLIGHEGQG SLLSYLKKKK WANSVYAGIV EDYDEFSLFV VSFDVTEDGV ERADDVLKAM | 720 |
| FQYMHLMRAS PWDKWVFDEL EIMSKTHFMF QSKSPPADFT SVVAANMHTF PKRDIVSEGV | 780 |
| LYFPHEWEQA LELLRLMTPE QMRVLIACQT LEERAVSEEK WYGTKYREMP LPIKFLEEMA | 840 |
| NPGANCALRL PHPNDFVVTD LNLVDERTVD TQHNHPHTIR NDDFCRVWYK PDVTFKKPRT | 900 |
| FAVATFHSPE VNPTPYSYAL SALFVSCLKD ELNEYSYDAL LAGMNYKLRL NGSNIYLSAG | 960 |
| GYSSKLPILV QRILEVMGSF ETHIGDEAFK RVKHAKCRSF ENMRLEEAHR HAVQQESNLL | 1020 |
| HERSWDIDEI VSAIRSCSFR DVIAHSKRLF RQVYCDILLY GNLKRSEAMD LADLIVDQVR | 1080 |
| APRALSMPST SKYWIGRQVQ LSCGVHYIYK CVHPNPDNAN CAVNCIYQIG VENYIDRAKL | 1140 |
| ALFSQMVDEP LFDQLRTKEQ LGYTVYSTPS RANDVQSFKV VVQSNVAPPE LIEQRIEAFW | 1200 |
| VEFRKTVADT SADQLQKHIQ SVVKGYIEKP KSQEEEVQAL LVEVANHQYE FGRKTKLAKL | 1260 |
| VRTLQLSDVL QFFDDYIRPE GPMRKKLSVH IYGNETRLEK LGDCSESGWS AFDNQSQTGL | 1320 |
| MAAMALASAG PASRDVKTEY IKDSQDFKRR TLVYEAPVAA L | 1361 |
Enzyme Prediction help
| EC | 3.2.1.8:31 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 37 | 335 | 2.7e-83 | 0.9438943894389439 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223956 | Ptr | 0.0 | 377 | 1317 | 13 | 928 | Secreted/periplasmic Zn-dependent peptidases, insulinase-like [Posttranslational modification, protein turnover, chaperones]. |
| 185056 | PRK15101 | 3.16e-98 | 378 | 1281 | 34 | 918 | protease3; Provisional |
| 395262 | Glyco_hydro_10 | 1.04e-96 | 36 | 352 | 1 | 308 | Glycosyl hydrolase family 10. |
| 406571 | Peptidase_M16_M | 1.93e-88 | 748 | 1033 | 1 | 283 | Middle or third domain of peptidase_M16. Peptidase_M16_M is the third domain of peptidase_M16 in eukaryotes of the insulin-degrading-enzyme type. Insulin-degrading enzymes - insulysin - are zinc metallopeptidases that metabolize several bioactive peptides, including insulin and the amyloid-beta-peptide. The tertiary structure of insulin-degrading enzymes resembles a clamshell composed of four structurally similar domains arranged to enclose a large central chamber. Substrates must enter the chamber, and it is likely that a hinge-like conformational change allows substrate binding and product release. Triphosphates are found to dock between the inner surfaces of the non-catalytic domains three and four. |
| 214750 | Glyco_10 | 6.51e-80 | 81 | 352 | 1 | 263 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOV63057.1|GH10 | 4.02e-200 | 2 | 358 | 1 | 355 |
| AOV63056.1|GH10 | 9.80e-184 | 6 | 357 | 5 | 358 |
| UIZ28775.1|GH10 | 4.87e-160 | 1 | 357 | 1 | 355 |
| UIZ24493.1|GH10 | 4.29e-156 | 1 | 357 | 1 | 353 |
| AOV63055.1|GH10 | 1.54e-127 | 15 | 356 | 12 | 350 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B70_A | 1.60e-204 | 378 | 1349 | 9 | 955 | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin [Homo sapiens],6B70_B Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin [Homo sapiens],6B7Y_A Cryo-EM structure of human insulin degrading enzyme [Homo sapiens],6B7Y_B Cryo-EM structure of human insulin degrading enzyme [Homo sapiens],6B7Z_A Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain [Homo sapiens],6B7Z_B Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain [Homo sapiens],6BF6_A Cryo-EM structure of human insulin degrading enzyme [Homo sapiens],6BF6_B Cryo-EM structure of human insulin degrading enzyme [Homo sapiens],6BF7_A Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain [Homo sapiens],6BF7_B Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain [Homo sapiens],6BF8_A Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens],6BF8_B Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens],6BF9_A Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain [Homo sapiens],6BF9_B Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain [Homo sapiens],6BFC_A Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens],6BFC_B Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens] |
| 2G47_A | 1.66e-204 | 378 | 1349 | 25 | 971 | Chain A, Insulin-degrading enzyme [Homo sapiens],2G47_B Chain B, Insulin-degrading enzyme [Homo sapiens],2G48_A Chain A, Insulin-degrading enzyme [Homo sapiens],2G48_B Chain B, Insulin-degrading enzyme [Homo sapiens],2G49_A Chain A, Insulin-degrading enzyme [Homo sapiens],2G49_B Chain B, Insulin-degrading enzyme [Homo sapiens],2G54_A Chain A, Insulin-degrading enzyme [Homo sapiens],2G54_B Chain B, Insulin-degrading enzyme [Homo sapiens],2G56_A Chain A, Insulin-degrading enzyme [Homo sapiens],2G56_B Chain B, Insulin-degrading enzyme [Homo sapiens],2JBU_A Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. [Homo sapiens],2JBU_B Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. [Homo sapiens],3E50_A Chain A, Insulin-degrading enzyme [Homo sapiens],3E50_B Chain B, Insulin-degrading enzyme [Homo sapiens] |
| 2JG4_A | 2.33e-204 | 378 | 1349 | 25 | 971 | Substrate-free IDE structure in its closed conformation [Homo sapiens],2JG4_B Substrate-free IDE structure in its closed conformation [Homo sapiens] |
| 3E4Z_A | 6.46e-204 | 378 | 1349 | 25 | 971 | Chain A, Insulin-degrading enzyme [Homo sapiens],3E4Z_B Chain B, Insulin-degrading enzyme [Homo sapiens] |
| 3QZ2_A | 1.27e-203 | 378 | 1295 | 25 | 933 | The structure of cysteine-free human insulin degrading enzyme [Homo sapiens],3QZ2_B The structure of cysteine-free human insulin degrading enzyme [Homo sapiens],4DTT_A Crystal structure of human insulin degrading enzyme (ide) in complex with compund 41367 [Homo sapiens],4DTT_B Crystal structure of human insulin degrading enzyme (ide) in complex with compund 41367 [Homo sapiens],4IFH_A Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 [Homo sapiens],4IFH_B Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 [Homo sapiens],4IOF_A Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) [Homo sapiens],4IOF_B Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) [Homo sapiens],4RE9_A Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 [Homo sapiens],4RE9_B Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 [Homo sapiens],5UOE_A Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE) [Homo sapiens],5UOE_B Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE) [Homo sapiens],5UOE_C Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE) [Homo sapiens],5UOE_D Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE) [Homo sapiens],5UOE_E Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE) [Homo sapiens],6B3Q_A Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens],6B3Q_B Cryo-EM structure of human insulin degrading enzyme in complex with insulin [Homo sapiens],7K1D_A Chain A, Insulin-degrading enzyme [Homo sapiens],7K1D_B Chain B, Insulin-degrading enzyme [Homo sapiens],7K1E_A Chain A, Insulin-degrading enzyme [Homo sapiens],7K1E_B Chain B, Insulin-degrading enzyme [Homo sapiens],7K1F_A Chain A, Insulin-degrading enzyme [Homo sapiens],7K1F_B Chain B, Insulin-degrading enzyme [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q24K02|IDE_BOVIN | 5.16e-204 | 384 | 1349 | 60 | 1000 | Insulin-degrading enzyme OS=Bos taurus OX=9913 GN=IDE PE=2 SV=1 |
| sp|P14735|IDE_HUMAN | 7.25e-204 | 378 | 1349 | 54 | 1000 | Insulin-degrading enzyme OS=Homo sapiens OX=9606 GN=IDE PE=1 SV=4 |
| sp|P35559|IDE_RAT | 3.03e-202 | 376 | 1349 | 52 | 1000 | Insulin-degrading enzyme OS=Rattus norvegicus OX=10116 GN=Ide PE=1 SV=1 |
| sp|Q9JHR7|IDE_MOUSE | 6.90e-200 | 376 | 1349 | 52 | 1000 | Insulin-degrading enzyme OS=Mus musculus OX=10090 GN=Ide PE=1 SV=1 |
| sp|Q06010|STE23_YEAST | 3.56e-182 | 383 | 1291 | 69 | 965 | A-factor-processing enzyme OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=STE23 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
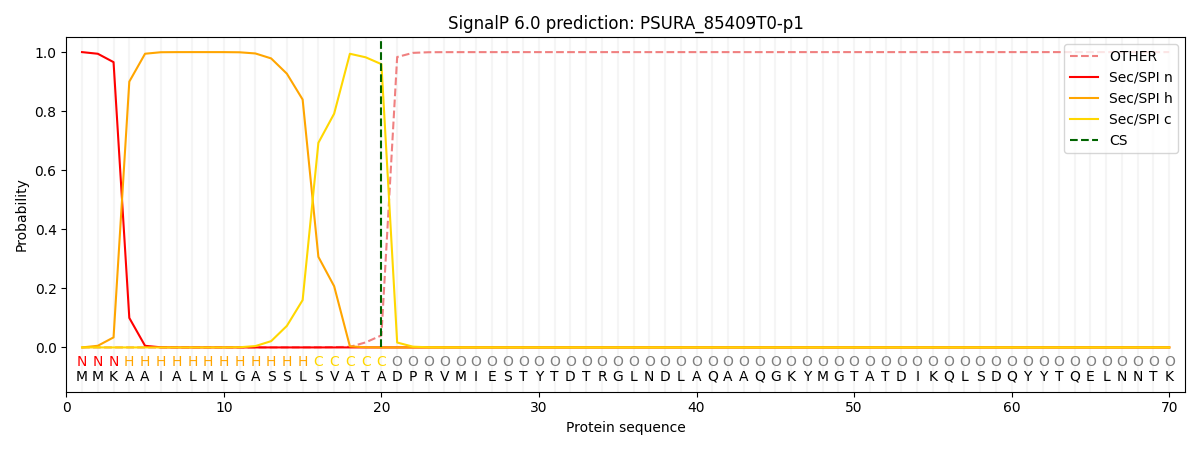
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000253 | 0.999714 | CS pos: 20-21. Pr: 0.9592 |
