You are browsing environment: FUNGIDB
CAZyme Information: PSURA_83548T0-p1
You are here: Home > Sequence: PSURA_83548T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
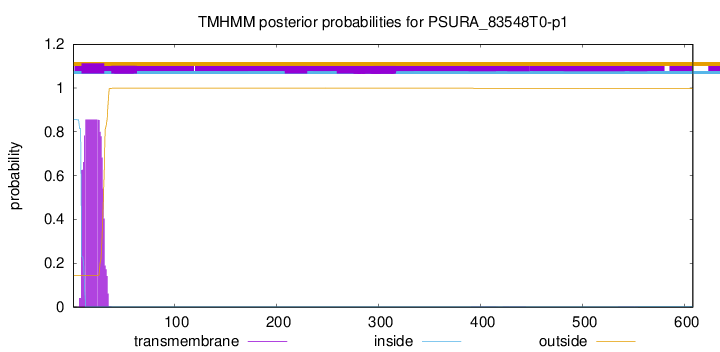
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_83548T0-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.122:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 237 | 446 | 3e-33 | 0.9791666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 2.28e-21 | 237 | 446 | 3 | 194 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 215596 | PLN03133 | 3.99e-13 | 220 | 489 | 385 | 634 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.05e-13 | 220 | 487 | 386 | 633 | |
| 3.38e-13 | 219 | 445 | 87 | 299 | |
| 3.46e-13 | 214 | 435 | 81 | 288 | |
| 4.85e-13 | 220 | 484 | 401 | 644 | |
| 7.76e-13 | 220 | 478 | 351 | 578 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.14e-14 | 214 | 435 | 81 | 288 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase B OS=Danio rerio OX=7955 GN=b3gnt5b PE=2 SV=1 |
|
| 1.95e-13 | 220 | 487 | 392 | 639 | Beta-1,3-galactosyltransferase GALT1 OS=Arabidopsis thaliana OX=3702 GN=GALT1 PE=1 SV=1 |
|
| 4.35e-13 | 222 | 487 | 373 | 617 | Hydroxyproline O-galactosyltransferase GALT3 OS=Arabidopsis thaliana OX=3702 GN=GALT3 PE=2 SV=1 |
|
| 5.97e-13 | 214 | 433 | 80 | 285 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Sus scrofa OX=9823 GN=B3GNT5 PE=2 SV=1 |
|
| 7.96e-13 | 214 | 471 | 81 | 328 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Xenopus tropicalis OX=8364 GN=b3gnt5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
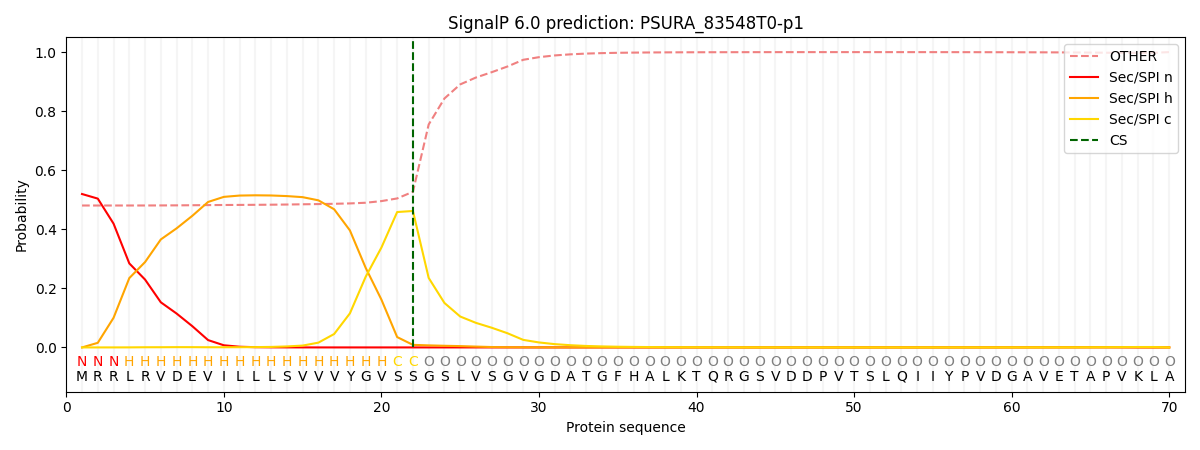
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.499322 | 0.500676 | CS pos: 22-23. Pr: 0.4620 |