You are browsing environment: FUNGIDB
CAZyme Information: PSURA_83139T0-p1
You are here: Home > Sequence: PSURA_83139T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_83139T0-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.151:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 91 | 239 | 3.2e-42 | 0.9871794871794872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 3.01e-41 | 35 | 240 | 3 | 207 | Glycosyl hydrolase family 12. |
| 235746 | PRK06215 | 2.43e-12 | 24 | 195 | 41 | 193 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.77e-177 | 1 | 242 | 1 | 242 | |
| 5.63e-147 | 1 | 242 | 1 | 243 | |
| 1.41e-140 | 5 | 240 | 7 | 242 | |
| 8.42e-122 | 9 | 242 | 11 | 243 | |
| 4.67e-118 | 1 | 239 | 1 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.71e-76 | 22 | 239 | 6 | 221 | Crystal structure of xeg-edgp [Aspergillus aculeatus],3VLB_D Crystal structure of xeg-edgp [Aspergillus aculeatus] |
|
| 5.91e-76 | 22 | 239 | 13 | 228 | Crystal structure of XEG [Aspergillus aculeatus],3VL9_A Crystal structure of xeg-xyloglucan [Aspergillus aculeatus],3VL9_B Crystal structure of xeg-xyloglucan [Aspergillus aculeatus] |
|
| 2.82e-72 | 22 | 239 | 26 | 242 | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus],4NPR_B Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus] |
|
| 3.68e-48 | 24 | 240 | 4 | 218 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
|
| 8.53e-47 | 24 | 240 | 5 | 219 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.60e-88 | 1 | 240 | 1 | 238 | Inactive glycoside hydrolase XLP1 OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=XLP1 PE=1 SV=1 |
|
| 1.93e-83 | 1 | 240 | 1 | 239 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
|
| 5.87e-83 | 1 | 240 | 1 | 241 | Xyloglucan-specific endo-beta-1,4-glucanase 1 OS=Phytophthora sojae (strain P6497) OX=1094619 GN=XEG1 PE=1 SV=1 |
|
| 1.18e-82 | 7 | 240 | 9 | 241 | Xyloglucan-specific endo-beta-1,4-glucanase 1 OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=XEG1 PE=1 SV=1 |
|
| 6.55e-82 | 22 | 239 | 23 | 239 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xgeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
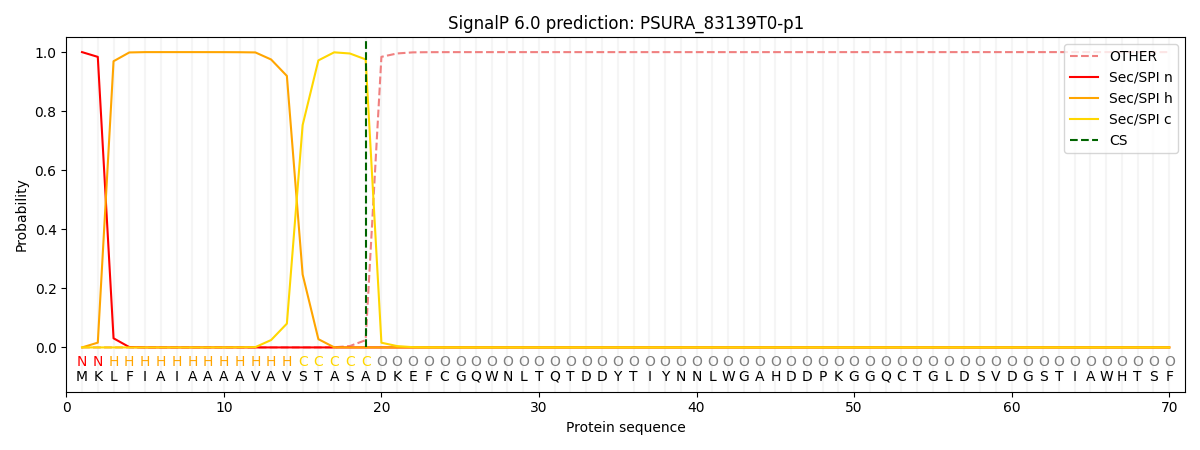
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000207 | 0.999746 | CS pos: 19-20. Pr: 0.9753 |
