You are browsing environment: FUNGIDB
CAZyme Information: PSURA_83132T0-p1
You are here: Home > Sequence: PSURA_83132T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_83132T0-p1 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:31 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 44 | 333 | 1.8e-73 | 0.9207920792079208 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 4.39e-89 | 35 | 350 | 1 | 308 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 1.12e-75 | 81 | 350 | 1 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 1.42e-61 | 33 | 334 | 21 | 330 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.21e-195 | 1 | 356 | 1 | 355 | |
| 8.09e-195 | 1 | 355 | 1 | 358 | |
| 3.40e-172 | 1 | 355 | 2 | 355 | |
| 3.97e-166 | 1 | 355 | 2 | 353 | |
| 1.21e-122 | 12 | 354 | 8 | 350 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.15e-78 | 34 | 348 | 2 | 327 | Chain A, Beta-xylanase [Bispora sp. MEY-1],5XZO_B Chain B, Beta-xylanase [Bispora sp. MEY-1],5XZU_A Chain A, Beta-xylanase [Bispora sp. MEY-1],5XZU_B Chain B, Beta-xylanase [Bispora sp. MEY-1] |
|
| 5.99e-74 | 34 | 353 | 2 | 316 | Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDT_B Crystal structure of GH10 family xylanase XynAF1 from Aspergillus fumigatus Z5 [Aspergillus fumigatus Z5],6JDY_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDY_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 120 minutes [Aspergillus fumigatus Z5],6JDZ_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JDZ_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 20 minutes [Aspergillus fumigatus Z5],6JE0_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE0_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 30 minutes [Aspergillus fumigatus Z5],6JE1_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE1_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 40 minutes [Aspergillus fumigatus Z5],6JE2_A Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5],6JE2_B Ligand complex structure of GH10 family xylanase XynAF1, soaking for 80 minutes [Aspergillus fumigatus Z5] |
|
| 9.88e-68 | 31 | 352 | 1 | 310 | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 3.10e-66 | 31 | 352 | 1 | 310 | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
|
| 1.46e-65 | 37 | 353 | 26 | 338 | GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8M_B GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8N_A GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872],6Q8N_B GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.86e-64 | 31 | 352 | 39 | 348 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
|
| 3.35e-60 | 35 | 354 | 19 | 331 | Endo-1,4-beta-xylanase OS=Agaricus bisporus OX=5341 GN=xlnA PE=2 SV=1 |
|
| 4.65e-59 | 39 | 321 | 24 | 302 | Endo-1,4-beta-xylanase D OS=Talaromyces funiculosus OX=28572 GN=xynD PE=1 SV=1 |
|
| 2.63e-57 | 30 | 328 | 33 | 328 | Endo-1,4-beta-xylanase 5 OS=Magnaporthe grisea OX=148305 GN=XYL5 PE=3 SV=1 |
|
| 2.63e-57 | 30 | 328 | 33 | 328 | Endo-1,4-beta-xylanase 5 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL5 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
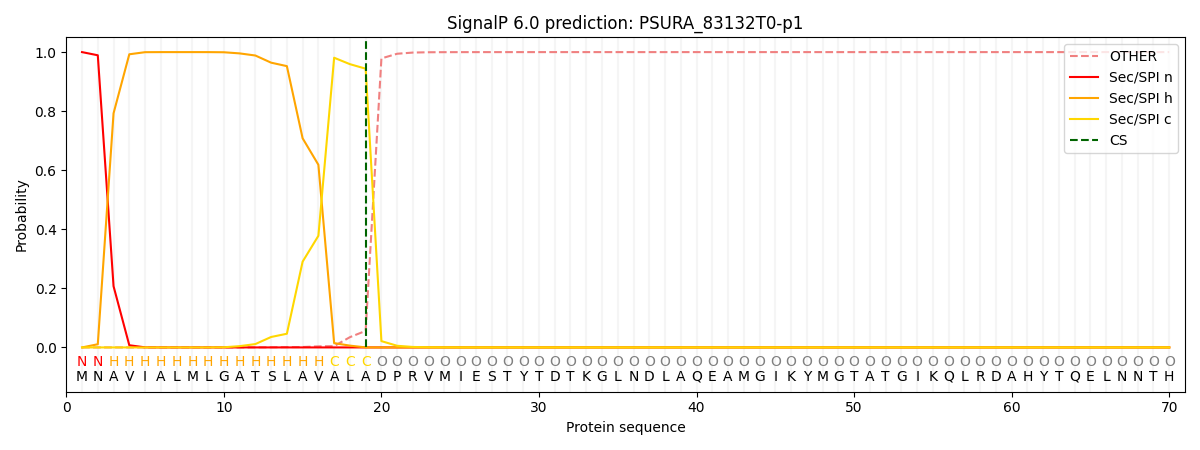
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000210 | 0.999772 | CS pos: 19-20. Pr: 0.9438 |
