You are browsing environment: FUNGIDB
CAZyme Information: PSURA_82591T0-p1
You are here: Home > Sequence: PSURA_82591T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_82591T0-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 132125; End:138847 Strand: + | |||||||||||
Full Sequence Download help
| MLQTLSSVAA LVVAASMVSG LPVPDGTWPV SKGSVKFEAV HLIKAGEVFD GKMQTFDRTN | 60 |
| ITCLGQEESG QSTAVFQLEA NATLKNAIIG KNQMEGVHCD DNDCTIDNVW WDDVCEDALS | 120 |
| IKGGDASSVS RVLGGGARYA DDKVIQHNGL GTVIIDGFYA QDFGKLYRSC GSCSNNPAQR | 180 |
| FLNMRNVFAD LEIIQAQRVD PNVSIVMMNE NYGDEAVLHN IYVKPGAENY TECGWSRGVQ | 240 |
| KGDKPLILGN GPKDPVCQYT MDEIHLTEGD RSMQQDQVVA TVHPAKEEKQ TALATMDKLV | 300 |
| QLQAALRAIE TGKRLESVDN VLVRQVLLQP RAQPQQATRE HDARDIYREL QALRKAKAKA | 360 |
| SAKQNVRDNE SSGETSDVGA TPPSPKKTLA PAHHHCSVPV VDGRLESAPS SPERHSNNDN | 420 |
| ELVLSTADTI ELMQKAFLTR EKELKRSEKA VARADAELRE ELSRAEKILP SKFLFERNLA | 480 |
| GDRALEAARN VLLRFQHRFY QLYFRQWWRA TLELRLQAQR RAVAEIVRVY RGHRGRQEAR | 540 |
| RLRRELSVLQ AQERQLLAFR MKFRSSQATK LQMAWRRYLR HRAIKYRQAR QAAARLLQQA | 600 |
| FRTRQWRGQS LVNALANARK LFAVVAVQKL YRGHRTRCKL AEAKRRQRQE ARVQLALLRN | 660 |
| MSKQARAAWM IERCGAGYLI AHRAIFPYAM RLRWHRAVYQ LRRQAAAECI ARAVCKWFGV | 720 |
| EVRREAARAK QVGEWLALVQ REHQRSRSAA VLIQKHLRRW VQQRKFLMAQ ARRKKLARKT | 780 |
| RLAEKARRLE QTPQRKSPGW LGSDRKPPVA TSPKRKPSRF MSNNSLSPQP ARLMSFLKPI | 840 |
| PLKDGNTKAA RKTNVEAAAL VQRNFRRYCA RKKARVRAWK HKARTVEARV LQRRKAATEI | 900 |
| QRRVRGIAGR TLARRRRAER LLKSPAKGSV AVETFSSVDV DVAVARATAG AKGALDFAAF | 960 |
| VRVIGLLGEI SLAQQAQSAS PYWGRYDGTE ARVLALLWRS LLPHSSMQPL VLEFEAYVSS | 1020 |
| ELSRHAACLQ RLFARQHNKL HGAAMLVEMR RILHNEALSR AAVVLQTQAR KLLARRELRR | 1080 |
| RMKETYEKFL DPTWGLPYWM NPRSGYSTWQ KPRLLGAEDV CCEPVPFPPP ERTLKASCNG | 1140 |
| KKNCDRCAEW VCYDCDEFFC LECFGEYHKS TPVFTGEEDS KEGNDPSLEK DENVDPNDNR | 1200 |
| ATAKREHELE RLQLSLELVT LSSGNAKRVG THLAVFVDVV PTNLIRGKAA VSSAPCHFKH | 1260 |
| LADVNEPNAS RKSKKLVIEL TWTKYVLGLY ELARSEAHEN FKRFGALKPR FFVLDASLLL | 1320 |
| VEERRLSIRA RFFGLLIDEY VTVGVPLPGL GLLHPGSNEI WTSEDLRGWI RNRQTIRLKK | 1380 |
| LSPQEAQPAE RALCAWRQLE RWGSAEIEAP EPESQGKDAG AQPPEATETS WLVDTHPKQA | 1440 |
| ITERMVPLAQ SYDPPLPKTK RRKAAGEEEE VVEDVPVAMF LVEFSLDPKR TVWINHSLAE | 1500 |
| RFWEWKRLKL LARSERTAQR QRAKDYQKQQ KDKWKQHQER NRQENGAIET EDAPPGVESS | 1560 |
| AAVDAVVEQP WAASNSAWTA DGAATDAYYN YNYYLSTDAS TAADGRTGYD GYSTSYPASY | 1620 |
| DYGYASTAPE TAPPVVAEEA TGLVDSENYY ANGDYGSGTS NWYGTEYSAY EMSGYSPAAT | 1680 |
| SSDNSGWASY SAESQLSIDP NAWYANTGDA SYNWQQSQPA QDQGSGTSSH DSYSIDLPSA | 1740 |
| TNGYYDPYTD DQQYSTPQSE WEEVFDPATQ QTYYVNRVTN ETAWQLP | 1787 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 31 | 226 | 1.2e-75 | 0.9484536082474226 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 8.11e-85 | 26 | 243 | 2 | 200 | Pectate lyase. |
| 380815 | Bbox1 | 1.19e-05 | 1144 | 1174 | 2 | 36 | B-box-type 1 zinc finger (Bbox1). The B-box-type zinc finger is a short zinc binding domain of around 40 amino acid residues in length. It has been found in transcription factors, ribonucleoproteins and proto-oncoproteins, such as in TRIM (tripartite motif) proteins that consist of an N-terminal RING finger (originally called an A-box), followed by 1-2 B-box domains and a coiled-coil domain (also called RBCC for Ring, B-box, Coiled-Coil). The B-box-type zinc finger often presents in combination with other motifs, like RING zinc finger, NHL motif, coiled-coil or RFP domain, in functionally unrelated proteins, most likely mediating protein-protein interactions. Based on different consensus sequences and the spacing of the 7-8 zinc-binding residues, the B-box-type zinc fingers can be divided into two groups, type 1 (Bbox1: C6H2) and type 2 (Bbox2: CHC3H2). This family corresponds to the type 1 B-box (Bbox1). |
| 400658 | Sfi1 | 5.47e-04 | 482 | 779 | 240 | 561 | Sfi1 spindle body protein. This is a family of fungal spindle pole body proteins that play a role in spindle body duplication. They contain binding sites for calmodulin-like proteins called centrins which are present in microtubule-organising centers. |
| 224208 | YccC | 0.001 | 502 | 668 | 497 | 661 | Uncharacterized membrane protein YccC [Function unknown]. |
| 380909 | Bbox1_DUF2009 | 0.002 | 1143 | 1170 | 4 | 32 | B-box-type 1 zinc finger found in DUF2009 domain-containing proteins and similar proteins. This group is composed of uncharacterized proteins containing a zinc finger B-box domain and a DUF2009 domain, and similar zinc finger B-box domain-containing proteins. The B-box motif shows high sequence similarity with B-Box-type 1 zinc finger found in tripartite motif-containing proteins (TRIMs). The type 1 B-box (Bbox1) zinc finger is characterized by a C6H2 zinc-binding consensus motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADN18839.1|PL3_2 | 2.38e-118 | 3 | 283 | 3 | 292 |
| AHX74070.1|PL3_2 | 4.49e-118 | 3 | 283 | 3 | 292 |
| CCD28273.1|PL3_2 | 3.99e-102 | 17 | 209 | 2 | 194 |
| ADN18841.1|PL3_2 | 2.70e-91 | 6 | 266 | 7 | 252 |
| ADN18837.1|PL3_2 | 2.55e-84 | 5 | 264 | 12 | 258 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EE6_A | 1.53e-22 | 43 | 171 | 11 | 135 | Crystal Structure Of Pectate Lyase From Bacillus Sp. Strain Ksm-P15. [Bacillus sp. KSM-P15] |
| 3B90_A | 3.16e-22 | 55 | 242 | 8 | 200 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B90_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
| 3B4N_A | 3.29e-21 | 55 | 242 | 126 | 318 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B4N_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_A Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
| 4U49_A | 9.61e-19 | 12 | 242 | 99 | 321 | Chain A, Pectate lyase [Pectobacterium carotovorum],4U49_B Chain B, Pectate lyase [Pectobacterium carotovorum],4U4B_A Chain A, Pectate lyase [Pectobacterium carotovorum] |
| 4EW9_A | 1.46e-16 | 42 | 172 | 15 | 139 | The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725],4EW9_B The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|A1DCY5|PLYD_NEOFI | 2.78e-53 | 14 | 260 | 12 | 235 | Probable pectate lyase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyD PE=3 SV=1 |
| sp|Q5BA88|PLYD_EMENI | 7.65e-53 | 29 | 260 | 43 | 258 | Probable pectate lyase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyD PE=3 SV=1 |
| sp|B0YB89|PLYD_ASPFC | 2.16e-51 | 10 | 260 | 9 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyD PE=3 SV=1 |
| sp|Q4WGV9|PLYD_ASPFU | 2.16e-51 | 10 | 260 | 9 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyD PE=3 SV=1 |
| sp|Q5ATC7|PLYH_EMENI | 2.41e-48 | 29 | 260 | 41 | 254 | Pectate lyase H OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
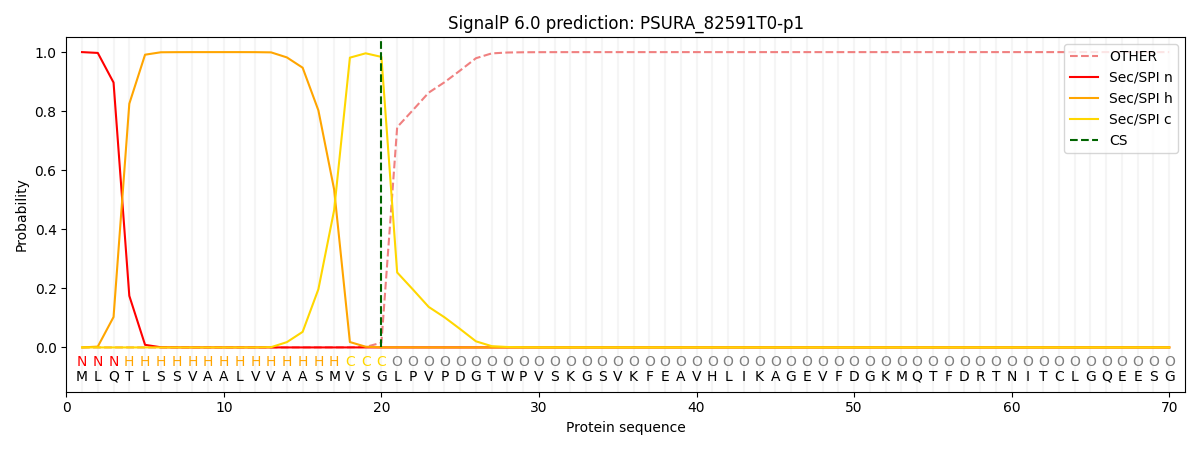
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000190 | 0.999770 | CS pos: 20-21. Pr: 0.9838 |
