You are browsing environment: FUNGIDB
CAZyme Information: PSURA_82055T0-p1
You are here: Home > Sequence: PSURA_82055T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
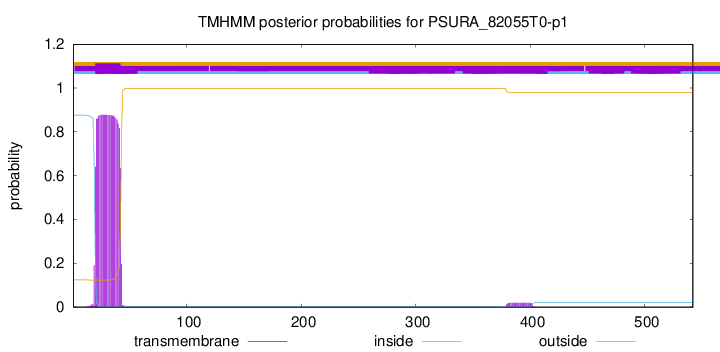
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_82055T0-p1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 157 | 424 | 1.9e-47 | 0.9924242424242424 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 1.33e-30 | 156 | 424 | 1 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
| 236940 | PRK11633 | 5.24e-09 | 83 | 150 | 66 | 143 | cell division protein DedD; Provisional |
| 223880 | TonB | 1.34e-06 | 94 | 161 | 60 | 128 | Periplasmic protein TonB, links inner and outer membranes [Cell wall/membrane/envelope biogenesis]. |
| 282710 | PT | 5.59e-06 | 110 | 144 | 2 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| 225805 | DamX | 2.11e-05 | 82 | 148 | 116 | 189 | Cell division protein DamX, binds to the septal ring, contains C-terminal SPOR domain [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.22e-111 | 148 | 524 | 45 | 431 | |
| 2.96e-110 | 156 | 523 | 131 | 515 | |
| 1.72e-86 | 156 | 436 | 65 | 355 | |
| 5.21e-66 | 156 | 494 | 64 | 435 | |
| 1.26e-63 | 156 | 508 | 91 | 453 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.15e-17 | 154 | 437 | 195 | 482 | Probable alpha-1,3-mannosyltransferase MNT4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT4 PE=3 SV=1 |
|
| 9.76e-15 | 156 | 390 | 275 | 514 | Putative alpha-1,3-mannosyltransferase MNN14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN14 PE=2 SV=1 |
|
| 1.05e-14 | 155 | 437 | 173 | 460 | Alpha-1,3-mannosyltransferase MNT2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT2 PE=1 SV=1 |
|
| 5.27e-13 | 156 | 436 | 264 | 575 | Putative alpha-1,3-mannosyltransferase MNN15 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN15 PE=2 SV=2 |
|
| 6.97e-13 | 156 | 436 | 331 | 635 | Putative alpha-1,3-mannosyltransferase MNN13 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN13 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
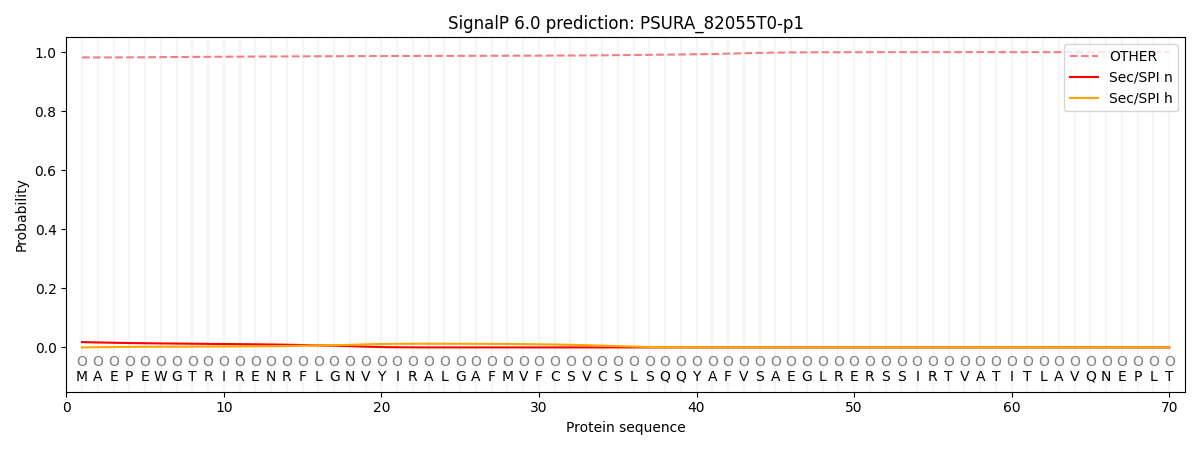
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.982617 | 0.017398 |