You are browsing environment: FUNGIDB
CAZyme Information: PSURA_80481T0-p1
You are here: Home > Sequence: PSURA_80481T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_80481T0-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 194 | 424 | 1.4e-50 | 0.9924242424242424 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238083 | PP2Cc | 4.20e-67 | 701 | 1015 | 1 | 252 | Serine/threonine phosphatases, family 2C, catalytic domain; The protein architecture and deduced catalytic mechanism of PP2C phosphatases are similar to the PP1, PP2A, PP2B family of protein Ser/Thr phosphatases, with which PP2C shares no sequence similarity. |
| 214625 | PP2Cc | 3.72e-63 | 698 | 1015 | 6 | 252 | Serine/threonine phosphatases, family 2C, catalytic domain. The protein architecture and deduced catalytic mechanism of PP2C phosphatases are similar to the PP1, PP2A, PP2B family of protein Ser/Thr phosphatases, with which PP2C shares no sequence similarity. |
| 395385 | PP2C | 5.42e-48 | 725 | 947 | 34 | 227 | Protein phosphatase 2C. Protein phosphatase 2C is a Mn++ or Mg++ dependent protein serine/threonine phosphatase. |
| 402574 | Mannosyl_trans3 | 7.69e-39 | 193 | 424 | 1 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
| 223704 | PTC1 | 1.71e-33 | 712 | 1015 | 20 | 249 | Serine/threonine protein phosphatase PrpC [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 5 | 565 | 589 | 1160 | |
| 9.31e-210 | 20 | 552 | 25 | 586 | |
| 3.86e-109 | 77 | 519 | 65 | 508 | |
| 7.17e-107 | 70 | 519 | 33 | 489 | |
| 1.10e-99 | 77 | 526 | 61 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.00e-30 | 727 | 1018 | 200 | 458 | Crystal structrual of a soluble protein [Oryza sativa Japonica Group] |
|
| 6.18e-30 | 723 | 1018 | 53 | 308 | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 [Arabidopsis thaliana],3NMN_D Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 [Arabidopsis thaliana] |
|
| 7.16e-30 | 723 | 1018 | 65 | 320 | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 [Arabidopsis thaliana] |
|
| 1.81e-29 | 730 | 1018 | 58 | 320 | Chain A, Probable protein phosphatase 2C 50 [Oryza sativa Japonica Group],5GWP_B Chain B, Probable protein phosphatase 2C 50 [Oryza sativa Japonica Group] |
|
| 1.85e-29 | 730 | 1018 | 59 | 321 | Chain A, Probable protein phosphatase 2C 50 [Oryza sativa Japonica Group],5ZCL_B Chain B, Probable protein phosphatase 2C 50 [Oryza sativa Japonica Group] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.69e-33 | 698 | 1015 | 23 | 270 | Probable protein phosphatase 2C 45 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0659500 PE=2 SV=2 |
|
| 4.31e-33 | 696 | 959 | 117 | 352 | Probable protein phosphatase 2C 11 OS=Arabidopsis thaliana OX=3702 GN=At1g43900 PE=2 SV=1 |
|
| 6.64e-33 | 698 | 1015 | 78 | 326 | Probable protein phosphatase 2C 10 OS=Oryza sativa subsp. japonica OX=39947 GN=Os02g0149800 PE=2 SV=1 |
|
| 1.92e-32 | 698 | 1015 | 60 | 308 | Probable protein phosphatase 2C 59 OS=Oryza sativa subsp. japonica OX=39947 GN=Os06g0698300 PE=2 SV=1 |
|
| 3.07e-32 | 698 | 1015 | 29 | 277 | Probable protein phosphatase 2C 71 OS=Arabidopsis thaliana OX=3702 GN=At5g24940 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
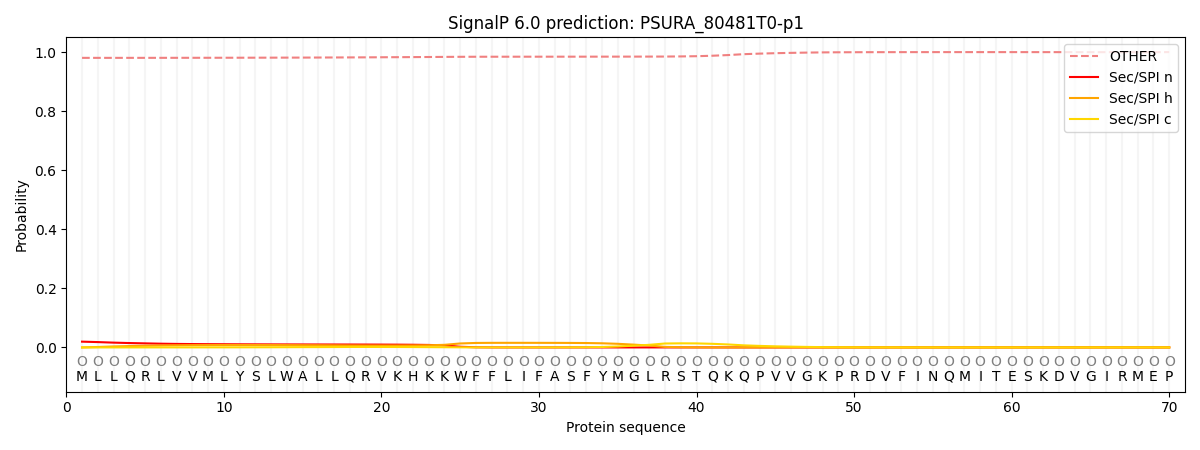
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.981833 | 0.018192 |
