You are browsing environment: FUNGIDB
CAZyme Information: PSURA_78827T0-p1
You are here: Home > Sequence: PSURA_78827T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
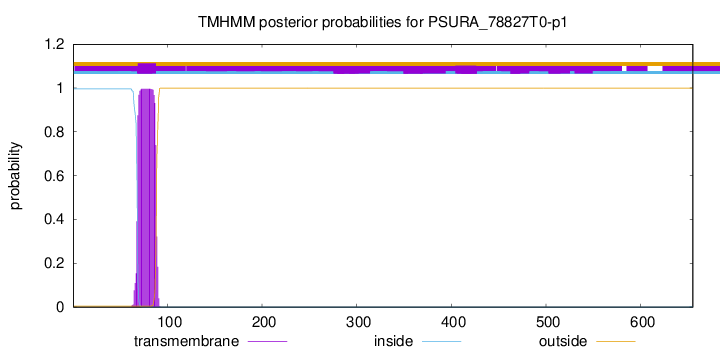
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_78827T0-p1 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 154 | 652 | 4.3e-24 | 0.9553072625698324 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 259922 | CuRO_1_Tth-MCO_like | 5.56e-60 | 129 | 265 | 1 | 139 | The first cupredoxin domain of the bacterial laccases similar to Tth-MCO from Thermus Thermophilus. The subfamily of bacterial laccases includes Tth-MCO and similar proteins. Tth-MCO is a hyperthermophilic multicopper oxidase (MCO) from thermus thermophilus HB27. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism in fungi and plants. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 259967 | CuRO_3_Tth-MCO_like | 1.11e-32 | 527 | 654 | 5 | 119 | The third cupredoxin domain of the bacterial laccases similar to Tth-MCO from Thermus Thermophilus. The subfamily of bacterial laccases includes Tth-MCO and similar proteins. Tth-MCO is a hyperthermophilic multicopper oxidase (MCO) from thermus thermophilus HB27. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism in fungi and plants. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 225043 | SufI | 1.79e-29 | 150 | 654 | 54 | 444 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 259870 | CuRO_3_LCC_like | 6.58e-29 | 527 | 654 | 3 | 129 | Cupredoxin domain 3 of laccase-like multicopper oxidases; including laccase, CueO, spore coat protein A, ascorbate oxidase and similar proteins. Laccase-like multicopper oxidases (MCOs) in this family contain three cupredoxin domains. They are able to couple oxidation of substrates with reduction of dioxygen to water. MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. Although the members of this family have diverse functions, majority of them have three cupredoxin domain repeats. The copper ions are bound in several sites; Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. Also included in this family are cupredoxin domains 2, 4, and 6 of the 6-domain MCO ceruloplasmin and similar proteins. |
| 400194 | Cu-oxidase_2 | 2.27e-20 | 522 | 652 | 4 | 128 | Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognized by the pfam00394 model. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.15e-299 | 191 | 654 | 1 | 464 | |
| 3.64e-26 | 150 | 654 | 10 | 471 | |
| 3.34e-11 | 245 | 654 | 67 | 474 | |
| 6.60e-11 | 136 | 651 | 26 | 481 | |
| 1.14e-10 | 158 | 653 | 167 | 662 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.56e-20 | 112 | 653 | 33 | 474 | Multicopper oxidase from Campylobacter jejuni: a metallo-oxidase [Campylobacter jejuni subsp. jejuni] |
|
| 2.12e-12 | 115 | 653 | 4 | 483 | Multicopper Oxidase CueO mutant E506A [Escherichia coli K-12] |
|
| 2.79e-12 | 115 | 653 | 4 | 483 | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis [Escherichia coli],1N68_A Copper bound to the Multicopper Oxidase CueO [Escherichia coli],2FQD_A Crystal Structures of E. coli Laccase CueO under different copper binding situations [Escherichia coli],2FQE_A Crystal Structures of E. coli Laccase CueO under different copper binding situations [Escherichia coli],2FQF_A Crystal Structures of E. coli Laccase CueO under different copper binding situations [Escherichia coli],2FQG_A Crystal Structures of E. coli Laccase CueO under different copper binding situations [Escherichia coli],3NSD_A Silver bound to the multicopper oxidase CueO (untagged) [Escherichia coli],3OD3_A CueO at 1.1 A resolution including residues in previously disordered region [Escherichia coli],3PAU_A CueO in the resting oxidized state [Escherichia coli str. K-12 substr. W3110],3PAV_A The reduced form of CueO [Escherichia coli] |
|
| 2.80e-12 | 115 | 653 | 4 | 483 | Multicopper Oxidase CueO (data2) [Escherichia coli K-12],4E9R_A Multicopper Oxidase CueO (data4) [Escherichia coli K-12],4E9S_A Multicopper Oxidase CueO (data5) [Escherichia coli K-12],4E9T_A Multicopper Oxidase CueO (data6) [Escherichia coli K-12],4EF3_A Multicopper Oxidase CueO (Citrate buffer) [Escherichia coli K-12],4NER_A Multicopper Oxidase CueO (data1) [Escherichia coli K-12] |
|
| 2.82e-12 | 115 | 653 | 8 | 487 | Structure of CueO - the signal peptide was truncated by HRV3C protease [Escherichia coli K-12],5B7M_A Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease [Escherichia coli K-12],5B7M_B Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease [Escherichia coli K-12],5B7M_C Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.54e-11 | 115 | 653 | 32 | 511 | Multicopper oxidase CueO OS=Escherichia coli (strain K12) OX=83333 GN=cueO PE=1 SV=2 |
|
| 2.79e-10 | 150 | 653 | 72 | 440 | Multicopper oxidase mco OS=Staphylococcus aureus OX=1280 GN=mco PE=1 SV=2 |
|
| 3.70e-10 | 150 | 653 | 72 | 440 | Multicopper oxidase mco OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=mco PE=3 SV=2 |
|
| 4.39e-10 | 115 | 653 | 32 | 511 | Multicopper oxidase CueO OS=Escherichia coli O157:H7 OX=83334 GN=cueO PE=3 SV=1 |
|
| 2.49e-08 | 121 | 653 | 42 | 440 | Multicopper oxidase mco OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=mco PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
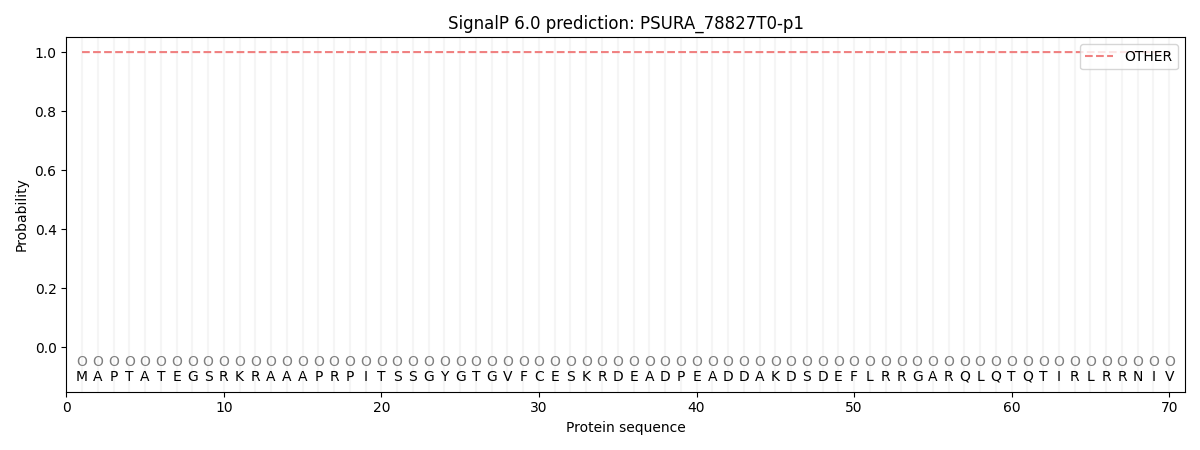
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000052 | 0.000004 |