You are browsing environment: FUNGIDB
CAZyme Information: PSURA_78175T0-p1
You are here: Home > Sequence: PSURA_78175T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_78175T0-p1 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 15 | 278 | 3e-75 | 0.9183673469387755 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237030 | kgd | 2.04e-07 | 221 | 305 | 40 | 124 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 237030 | kgd | 3.44e-05 | 247 | 307 | 44 | 104 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 236776 | PRK10856 | 6.41e-05 | 218 | 306 | 165 | 253 | cytoskeleton protein RodZ. |
| 237030 | kgd | 9.61e-05 | 226 | 306 | 34 | 114 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 411474 | fibronec_FbpA | 1.24e-04 | 218 | 305 | 153 | 233 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.65e-135 | 1 | 319 | 1 | 320 | |
| 9.48e-135 | 1 | 319 | 1 | 320 | |
| 5.45e-134 | 1 | 319 | 1 | 320 | |
| 7.73e-134 | 1 | 319 | 1 | 320 | |
| 7.73e-134 | 1 | 319 | 1 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.03e-11 | 93 | 195 | 63 | 170 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
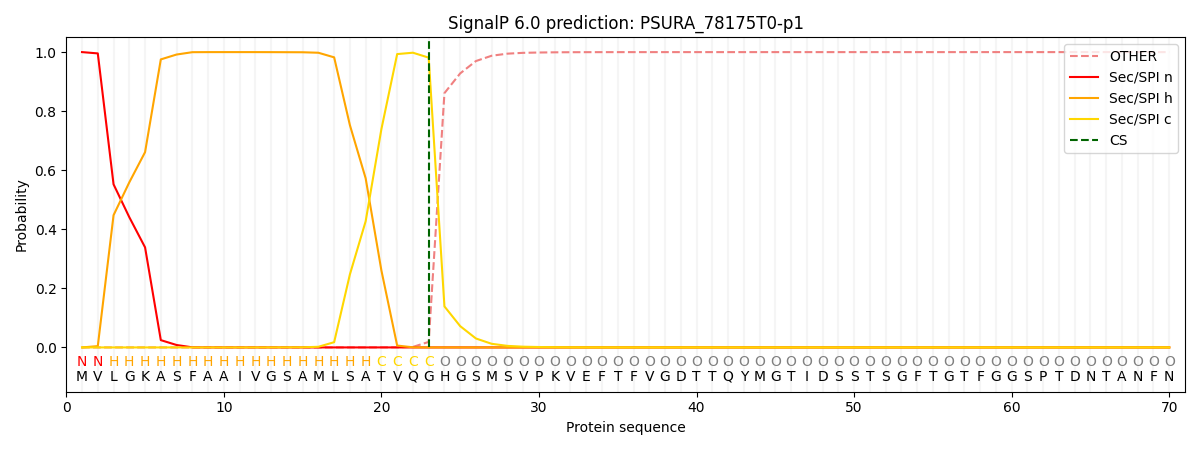
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999721 | CS pos: 23-24. Pr: 0.9813 |
