You are browsing environment: FUNGIDB
CAZyme Information: PSURA_74255T0-p1
You are here: Home > Sequence: PSURA_74255T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_74255T0-p1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 29 | 283 | 4.6e-20 | 0.977491961414791 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411408 | PspC_subgroup_2 | 2.33e-26 | 294 | 548 | 228 | 482 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 411408 | PspC_subgroup_2 | 2.93e-25 | 373 | 551 | 290 | 463 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 411408 | PspC_subgroup_2 | 5.44e-24 | 350 | 538 | 317 | 483 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 411408 | PspC_subgroup_2 | 3.32e-22 | 375 | 551 | 281 | 441 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 227625 | Scw11 | 8.77e-21 | 4 | 279 | 21 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.53e-119 | 20 | 323 | 565 | 870 | |
| 1.60e-117 | 1 | 287 | 1 | 288 | |
| 1.87e-72 | 1 | 287 | 1 | 288 | |
| 3.14e-54 | 1 | 199 | 1 | 200 | |
| 2.50e-46 | 29 | 287 | 22 | 282 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.92e-09 | 29 | 289 | 39 | 297 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.83e-10 | 56 | 230 | 340 | 516 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 4.83e-10 | 56 | 230 | 340 | 516 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 6.38e-10 | 56 | 233 | 340 | 519 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
|
| 8.28e-10 | 56 | 230 | 327 | 503 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 5.97e-09 | 70 | 286 | 68 | 303 | Glucan 1,3-beta-glucosidase BGL2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BGL2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
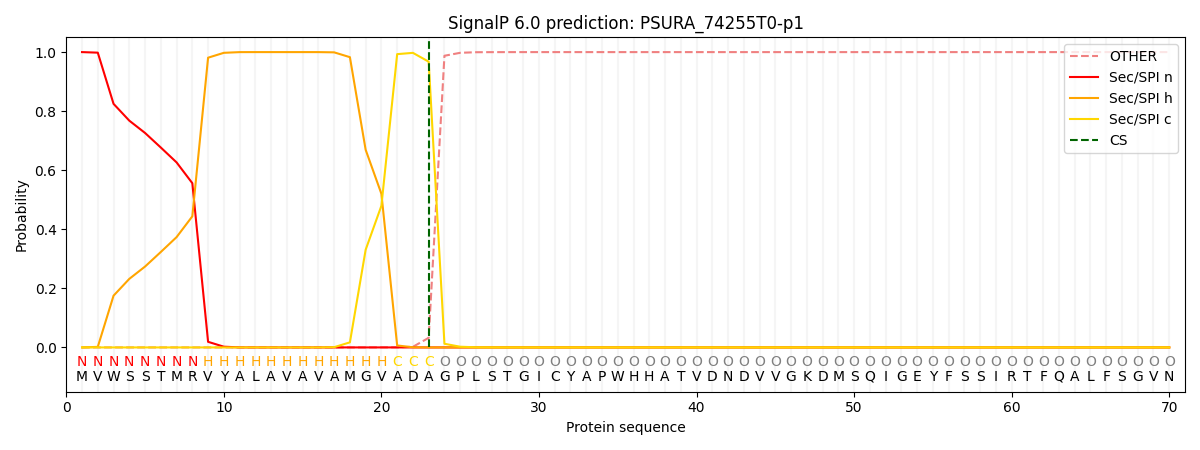
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000216 | 0.999773 | CS pos: 23-24. Pr: 0.9675 |
