You are browsing environment: FUNGIDB
CAZyme Information: PSURA_73616T0-p1
You are here: Home > Sequence: PSURA_73616T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
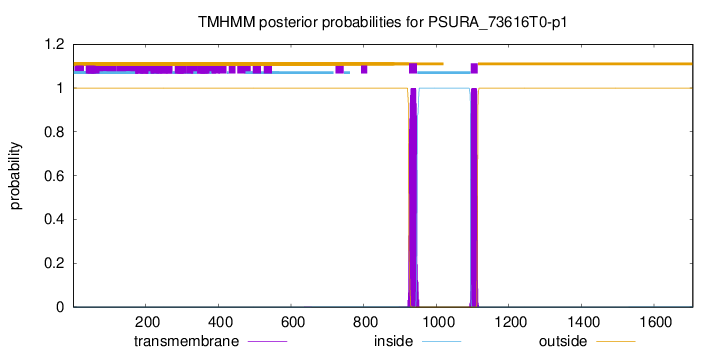
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_73616T0-p1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 1311 | 1544 | 1.1e-48 | 0.9924242424242424 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173791 | Peptidases_S8_Kp43_protease | 3.52e-82 | 375 | 689 | 3 | 293 | Peptidase S8 family domain in Kp43 proteases. Kp43 proteases are members of the peptidase S8 or Subtilase clan of proteases. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure (an example of convergent evolution). Kp43 is topologically similar to kexin and furin both of which are proprotein convertases, but differ in amino acids sequence and the position of its C-terminal barrel. Kp43 has 3 Ca2+ binding sites that differ from the corresponding sites in the other known subtilisin-like proteases. KP-43 protease is known to be an oxidation-resistant protease when compared with the other subtilisin-like proteases |
| 395035 | Peptidase_S8 | 2.25e-42 | 378 | 697 | 1 | 281 | Subtilase family. Subtilases are a family of serine proteases. They appear to have independently and convergently evolved an Asp/Ser/His catalytic triad, like that found in the trypsin serine proteases (see pfam00089). Structure is an alpha/beta fold containing a 7-stranded parallel beta sheet, order 2314567. |
| 402574 | Mannosyl_trans3 | 6.97e-38 | 1311 | 1544 | 2 | 273 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
| 173812 | Peptidases_S8_1 | 8.48e-36 | 378 | 654 | 1 | 245 | Peptidase S8 family domain, uncharacterized subfamily 1. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| 173787 | Peptidases_S8_S53 | 2.44e-31 | 381 | 654 | 1 | 224 | Peptidase domain in the S8 and S53 families. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) family include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53, it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium; some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.59e-114 | 1315 | 1705 | 1 | 364 | |
| 4.05e-91 | 1205 | 1703 | 106 | 575 | |
| 2.10e-88 | 1205 | 1565 | 98 | 466 | |
| 2.14e-88 | 1205 | 1696 | 143 | 596 | |
| 7.21e-82 | 1205 | 1561 | 72 | 421 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-38 | 375 | 845 | 18 | 404 | Chain A, Subtilase SubHal from Bacillus halmapalus [Sutcliffiella halmapala],5FAX_B Chain B, Subtilase SubHal from Bacillus halmapalus [Sutcliffiella halmapala],5FBZ_A Chain A, Enzyme subtilase SubHal from Bacillus halmapalus [Sutcliffiella halmapala],5FBZ_C Chain C, Enzyme subtilase SubHal from Bacillus halmapalus [Sutcliffiella halmapala] |
|
| 2.07e-37 | 365 | 845 | 7 | 405 | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) [Bacillus sp. KSM-KP43],1WME_A Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) [Bacillus sp. KSM-KP43] |
|
| 6.86e-37 | 365 | 845 | 7 | 405 | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) [Bacillus sp. KSM-KP43] |
|
| 6.18e-14 | 375 | 668 | 135 | 387 | Chain A, Subtilisin-like serine protease [Thermococcus kodakarensis],3AFG_B Chain B, Subtilisin-like serine protease [Thermococcus kodakarensis] |
|
| 1.80e-10 | 365 | 652 | 10 | 233 | Subtilisin Dy In Complex With The Synthetic Inhibitor N- Benzyloxycarbonyl-Ala-Pro-Phe-Chloromethyl Ketone [Bacillus licheniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-35 | 376 | 840 | 327 | 866 | Serine protease/ABC transporter B family protein tagD OS=Dictyostelium discoideum OX=44689 GN=tagD PE=3 SV=1 |
|
| 6.74e-34 | 375 | 813 | 300 | 764 | Serine protease/ABC transporter B family protein tagA OS=Dictyostelium discoideum OX=44689 GN=tagA PE=1 SV=2 |
|
| 1.57e-31 | 376 | 847 | 376 | 916 | Serine protease/ABC transporter B family protein tagB OS=Dictyostelium discoideum OX=44689 GN=tagB PE=3 SV=2 |
|
| 4.43e-31 | 376 | 843 | 314 | 864 | Serine protease/ABC transporter B family protein tagC OS=Dictyostelium discoideum OX=44689 GN=tagC PE=2 SV=2 |
|
| 1.32e-18 | 1284 | 1546 | 219 | 528 | Alpha-1,2-mannosyltransferase MNN21 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN21 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
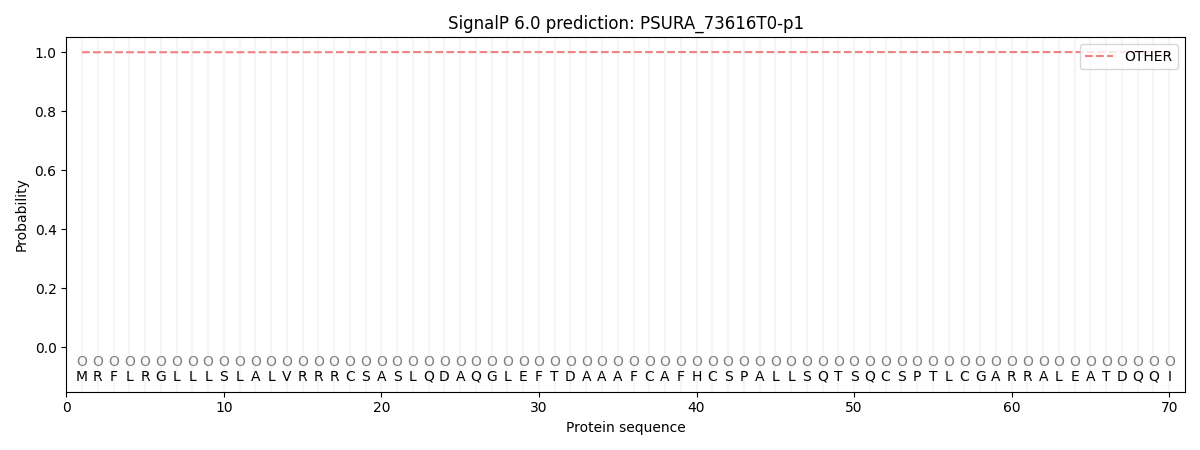
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999633 | 0.000412 |