You are browsing environment: FUNGIDB
CAZyme Information: PSURA_72148T0-p1
You are here: Home > Sequence: PSURA_72148T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_72148T0-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.55:24 | 3.2.1.37:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH54 | 21 | 329 | 7.8e-134 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 401229 | ArabFuran-catal | 0.0 | 21 | 329 | 2 | 315 | Alpha-L-arabinofuranosidase B, catalytic. Members of this family, which are present in fungal alpha-L-arabinofuranosidase B, adopt a beta-sandwich fold similar to that of Concanavalin A-like lectins/glucanase. The beta-sandwich fold consists of two anti-parallel beta-sheets with seven and and six strands, respectively. In addition, there are four helices outside of the beta-strands. The beta-sandwich strands are closely packed and curved with a jelly roll topology, creating a small catalytic pocket. The domain catalyzes the hydrolysis of alpha-1,2-, alpha-1,3- and alpha-1,5-L-arabinofuranosidic bonds in L-arabinose-containing hemicelluloses such as arabinoxylan and L-arabinan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.39e-165 | 1 | 328 | 1 | 327 | |
| 2.27e-131 | 16 | 332 | 18 | 335 | |
| 5.68e-131 | 16 | 332 | 80 | 397 | |
| 6.11e-131 | 21 | 328 | 176 | 488 | |
| 6.11e-131 | 21 | 328 | 176 | 488 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.11e-122 | 19 | 328 | 2 | 316 | Crystal structure of arabinofuranosidase [Aspergillus luchuensis],1WD4_A Crystal structure of arabinofuranosidase complexed with arabinose [Aspergillus luchuensis],6SXS_AAA Chain AAA, Alpha-L-arabinofuranosidase B [Aspergillus luchuensis] |
|
| 1.15e-122 | 19 | 328 | 3 | 317 | GH54 a-l-arabinofuranosidase soaked with aziridine inhibitor [Aspergillus luchuensis IFO 4308] |
|
| 3.16e-122 | 19 | 328 | 2 | 316 | E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside [Aspergillus luchuensis IFO 4308] |
|
| 8.97e-122 | 19 | 328 | 2 | 316 | Chain A, alpha-L-arabinofuranosidase B [Aspergillus luchuensis],2D44_A Chain A, alpha-L-arabinofuranosidase B [Aspergillus luchuensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.30e-124 | 7 | 332 | 7 | 337 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=abfB PE=3 SV=1 |
|
| 5.30e-124 | 7 | 332 | 7 | 337 | Alpha-L-arabinofuranosidase B OS=Aspergillus niger OX=5061 GN=abfB PE=1 SV=1 |
|
| 1.87e-123 | 3 | 329 | 5 | 342 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfB PE=3 SV=1 |
|
| 4.26e-123 | 7 | 328 | 7 | 333 | Alpha-L-arabinofuranosidase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfB PE=1 SV=1 |
|
| 6.02e-122 | 18 | 332 | 26 | 345 | Probable alpha-L-arabinofuranosidase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=abfB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
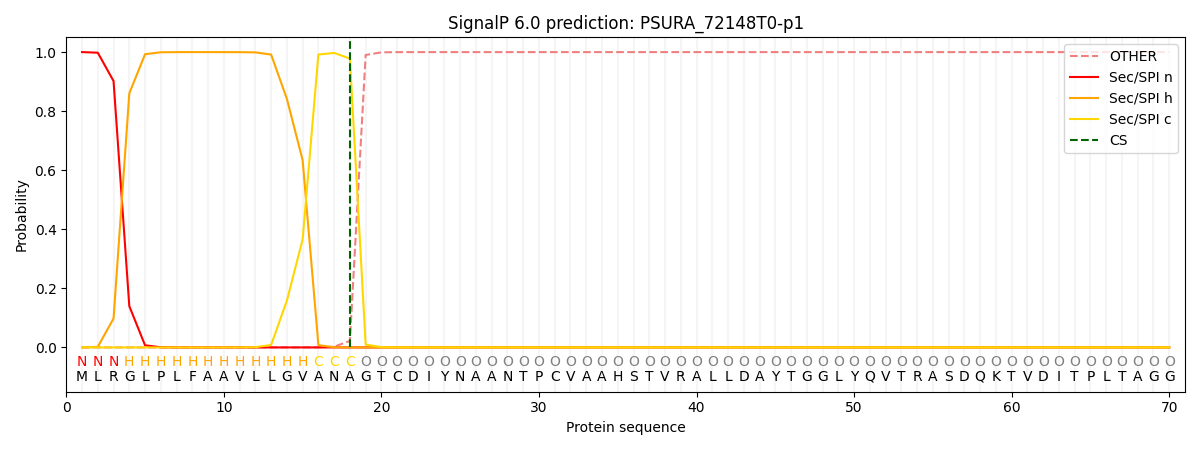
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000234 | 0.999751 | CS pos: 18-19. Pr: 0.9780 |
