You are browsing environment: FUNGIDB
CAZyme Information: PSURA_72080T0-p1
You are here: Home > Sequence: PSURA_72080T0-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora ramorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora ramorum | |||||||||||
| CAZyme ID | PSURA_72080T0-p1 | |||||||||||
| CAZy Family | AA6 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.151:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 91 | 239 | 4.5e-42 | 0.9871794871794872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 4.57e-42 | 32 | 239 | 1 | 206 | Glycosyl hydrolase family 12. |
| 235746 | PRK06215 | 9.29e-12 | 32 | 219 | 50 | 211 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.94e-176 | 1 | 242 | 1 | 242 | |
| 1.86e-140 | 1 | 241 | 1 | 240 | |
| 1.86e-140 | 1 | 241 | 1 | 240 | |
| 8.95e-116 | 1 | 239 | 1 | 239 | |
| 1.68e-115 | 6 | 240 | 7 | 240 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.82e-72 | 17 | 239 | 22 | 242 | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus],4NPR_B Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus [Aspergillus niveus] |
|
| 4.16e-71 | 14 | 240 | 6 | 229 | Crystal structure of XEG [Aspergillus aculeatus],3VL9_A Crystal structure of xeg-xyloglucan [Aspergillus aculeatus],3VL9_B Crystal structure of xeg-xyloglucan [Aspergillus aculeatus] |
|
| 9.46e-71 | 21 | 240 | 6 | 222 | Crystal structure of xeg-edgp [Aspergillus aculeatus],3VLB_D Crystal structure of xeg-edgp [Aspergillus aculeatus] |
|
| 5.40e-45 | 21 | 240 | 2 | 219 | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 [Aspergillus aculeatus],5GM3_B Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 [Aspergillus aculeatus] |
|
| 2.09e-44 | 20 | 239 | 1 | 217 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.11e-82 | 3 | 239 | 4 | 238 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
|
| 1.64e-79 | 1 | 239 | 1 | 237 | Inactive glycoside hydrolase XLP1 OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=XLP1 PE=1 SV=1 |
|
| 8.17e-75 | 1 | 239 | 1 | 237 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xgeA PE=3 SV=1 |
|
| 8.47e-75 | 1 | 239 | 1 | 249 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xgeA PE=3 SV=2 |
|
| 2.33e-74 | 1 | 239 | 1 | 237 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xgeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
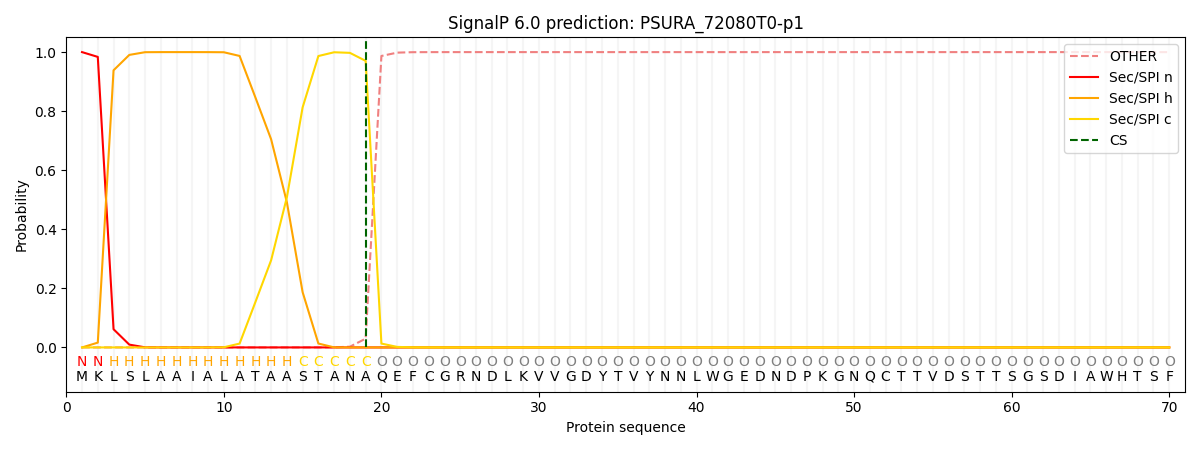
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000255 | 0.999720 | CS pos: 19-20. Pr: 0.9703 |
