You are browsing environment: FUNGIDB
CAZyme Information: PSK41430.1
You are here: Home > Sequence: PSK41430.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
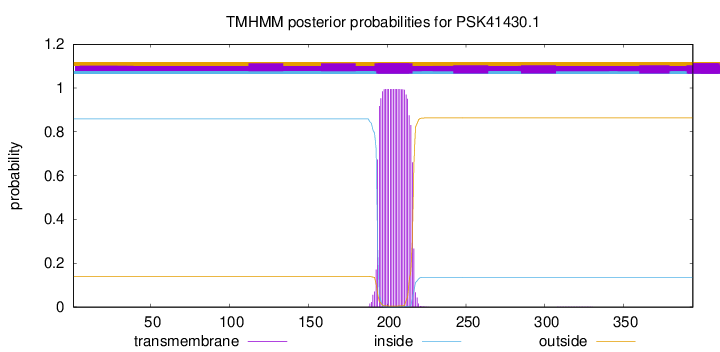
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK41430.1 | |||||||||||
| CAZy Family | GT91 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 4.35e-13 | 248 | 392 | 32 | 173 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.76e-201 | 25 | 392 | 64 | 429 | |
| 4.60e-38 | 125 | 392 | 113 | 382 | |
| 1.67e-37 | 125 | 392 | 113 | 382 | |
| 9.63e-25 | 175 | 382 | 189 | 400 | |
| 4.02e-23 | 205 | 392 | 193 | 385 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.36e-10 | 261 | 392 | 380 | 522 | Putative glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=btgC PE=3 SV=2 |
|
| 7.48e-10 | 239 | 381 | 323 | 473 | Glucan endo-1,3-beta-glucosidase btgC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=btgC PE=2 SV=1 |
|
| 2.08e-06 | 284 | 371 | 64 | 151 | Glucan 1,3-beta-glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=bgl2 PE=2 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
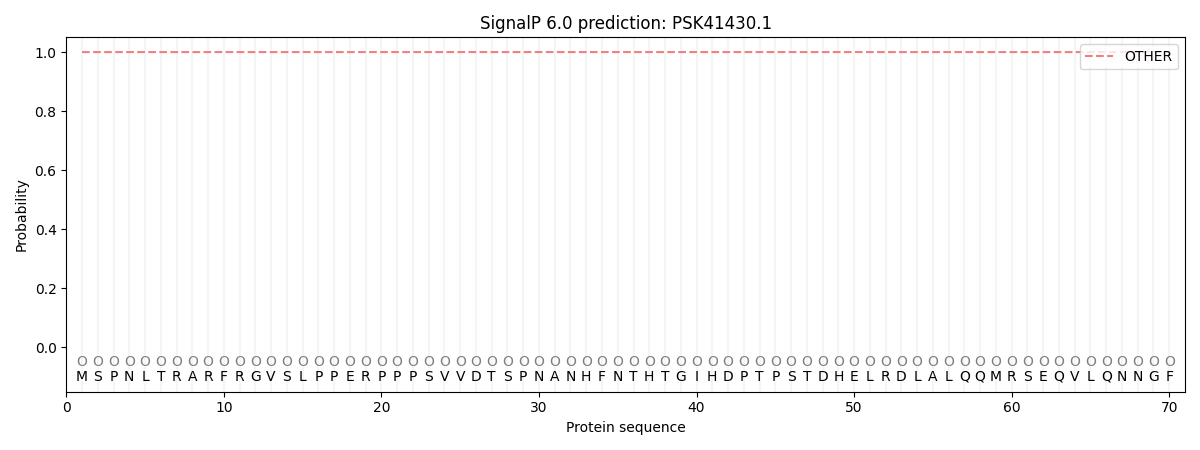
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000068 | 0.000000 |