You are browsing environment: FUNGIDB
CAZyme Information: PSK40580.1
You are here: Home > Sequence: PSK40580.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK40580.1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 10 | 404 | 2.2e-78 | 0.9459459459459459 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403214 | DUF3445 | 1.90e-94 | 488 | 726 | 1 | 231 | Protein of unknown function (DUF3445). This family of proteins are functionally uncharacterized. This protein is found in bacteria and eukaryotes. Proteins in this family are typically between 264 to 418 amino acids in length. This protein has a conserved RLP sequence motif. This protein has two completely conserved R residues that may be functionally important. |
| 282094 | Mannosyl_trans2 | 6.21e-42 | 67 | 419 | 44 | 424 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
| 227829 | COG5542 | 2.67e-22 | 79 | 385 | 77 | 364 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 789 | 1 | 798 | |
| 6.96e-180 | 2 | 412 | 3 | 416 | |
| 6.96e-180 | 2 | 401 | 3 | 402 | |
| 3.14e-132 | 10 | 401 | 11 | 410 | |
| 3.14e-132 | 10 | 401 | 11 | 410 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.24e-18 | 521 | 766 | 86 | 331 | Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) [Pseudomonas mendocina ymp],5LTH_A Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) in complex with the dimethylamine substrate [Pseudomonas mendocina],5LTI_A Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) in complex with the dimethylamine substrate [Pseudomonas mendocina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.70e-96 | 10 | 398 | 11 | 401 | GPI mannosyltransferase 2 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI18 PE=3 SV=1 |
|
| 1.84e-70 | 10 | 385 | 9 | 361 | GPI mannosyltransferase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI18 PE=3 SV=1 |
|
| 7.77e-52 | 10 | 399 | 11 | 401 | GPI mannosyltransferase 2 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=GPI18 PE=3 SV=1 |
|
| 5.87e-47 | 4 | 382 | 5 | 382 | GPI mannosyltransferase 2 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=GPI18 PE=3 SV=1 |
|
| 1.08e-45 | 75 | 391 | 56 | 385 | GPI mannosyltransferase 2 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=GPI18 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.846156 | 0.153847 |
TMHMM Annotations download full data without filtering help
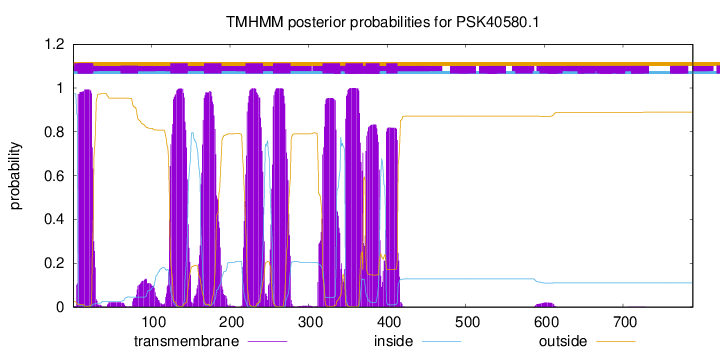
| Start | End |
|---|---|
| 7 | 26 |
| 124 | 146 |
| 167 | 189 |
| 220 | 242 |
| 255 | 277 |
| 318 | 340 |
| 347 | 369 |
| 373 | 390 |
| 399 | 413 |
