You are browsing environment: FUNGIDB
CAZyme Information: PSK39922.1
You are here: Home > Sequence: PSK39922.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK39922.1 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:29 | 3.2.1.21:6 | 3.2.1.75:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 106 | 427 | 1.7e-117 | 0.9966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 181856 | thrA | 1.14e-114 | 438 | 789 | 464 | 813 | bifunctional aspartokinase I/homoserine dehydrogenase I; Provisional |
| 223536 | ThrA | 3.99e-88 | 438 | 791 | 2 | 329 | Homoserine dehydrogenase [Amino acid transport and metabolism]. |
| 236530 | metL | 9.32e-73 | 434 | 789 | 453 | 807 | bifunctional aspartate kinase II/homoserine dehydrogenase II; Provisional |
| 395601 | Homoserine_dh | 7.31e-64 | 584 | 784 | 1 | 177 | Homoserine dehydrogenase. |
| 225344 | BglC | 1.07e-59 | 44 | 431 | 11 | 369 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 794 | 1 | 795 | |
| 0.0 | 127 | 794 | 1 | 671 | |
| 3.22e-248 | 21 | 431 | 31 | 440 | |
| 2.61e-247 | 21 | 431 | 31 | 440 | |
| 7.44e-247 | 29 | 431 | 37 | 440 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.06e-233 | 436 | 794 | 1 | 359 | Chain A, Homoserine dehydrogenase [[Candida] auris],7M92_B Chain B, Homoserine dehydrogenase [[Candida] auris] |
|
| 1.25e-146 | 437 | 792 | 2 | 358 | Homoserine Dehydrogenase From S. Cerevisiae Complex With Nad+ [Saccharomyces cerevisiae],1EBF_B Homoserine Dehydrogenase From S. Cerevisiae Complex With Nad+ [Saccharomyces cerevisiae],1EBU_A Homoserine Dehydrogenase Complex With Nad Analogue And L- Homoserine [Saccharomyces cerevisiae],1EBU_B Homoserine Dehydrogenase Complex With Nad Analogue And L- Homoserine [Saccharomyces cerevisiae],1EBU_C Homoserine Dehydrogenase Complex With Nad Analogue And L- Homoserine [Saccharomyces cerevisiae],1EBU_D Homoserine Dehydrogenase Complex With Nad Analogue And L- Homoserine [Saccharomyces cerevisiae],1TVE_A Homoserine Dehydrogenase in complex with 4-(4-hydroxy-3-isopropylphenylthio)-2-isopropylphenol [Saccharomyces cerevisiae],1TVE_B Homoserine Dehydrogenase in complex with 4-(4-hydroxy-3-isopropylphenylthio)-2-isopropylphenol [Saccharomyces cerevisiae] |
|
| 1.29e-146 | 437 | 792 | 3 | 359 | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline [Saccharomyces cerevisiae],1Q7G_B Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline [Saccharomyces cerevisiae] |
|
| 7.19e-118 | 47 | 431 | 5 | 366 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 5.60e-117 | 47 | 431 | 5 | 366 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.13e-156 | 48 | 431 | 39 | 427 | Glucan 1,3-beta-glucosidase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=EXG2 PE=1 SV=2 |
|
| 6.65e-146 | 437 | 792 | 3 | 359 | Homoserine dehydrogenase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HOM6 PE=1 SV=1 |
|
| 3.93e-123 | 52 | 431 | 43 | 441 | Glucan 1,3-beta-glucosidase 1 OS=Wickerhamomyces anomalus OX=4927 GN=EXG1 PE=3 SV=1 |
|
| 1.14e-116 | 52 | 431 | 49 | 412 | Glucan 1,3-beta-glucosidase OS=Lachancea kluyveri (strain ATCC 58438 / CBS 3082 / BCRC 21498 / NBRC 1685 / JCM 7257 / NCYC 543 / NRRL Y-12651) OX=226302 GN=EXG1 PE=3 SV=1 |
|
| 2.18e-116 | 47 | 431 | 49 | 410 | Glucan 1,3-beta-glucosidase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=XOG1 PE=1 SV=5 |
SignalP and Lipop Annotations help
This protein is predicted as SP
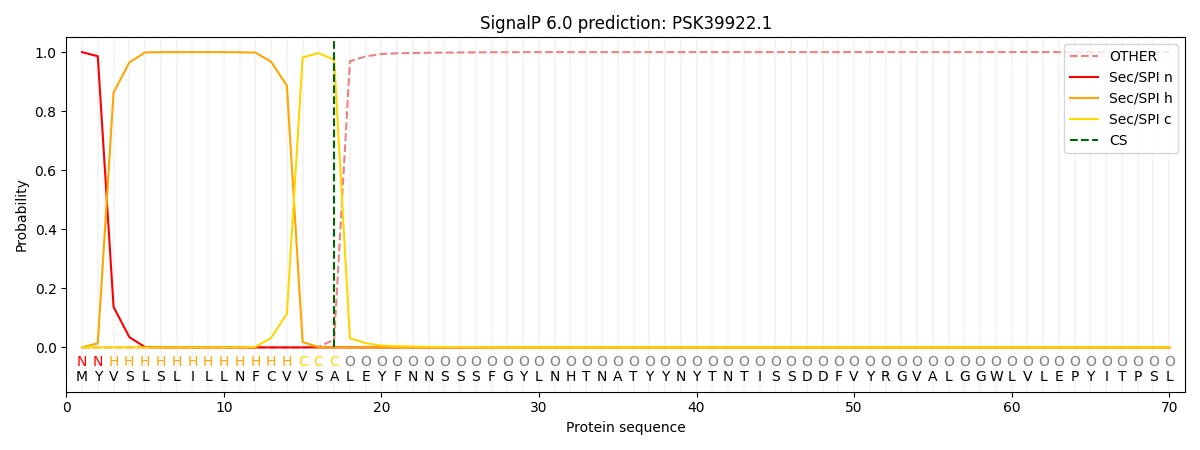
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000307 | 0.999637 | CS pos: 17-18. Pr: 0.9739 |
