You are browsing environment: FUNGIDB
CAZyme Information: PSK39837.1
You are here: Home > Sequence: PSK39837.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK39837.1 | |||||||||||
| CAZy Family | GT21 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 625909; End:630593 Strand: + | |||||||||||
Full Sequence Download help
| MGTSTVATTE EPLLTQNDTD GSEPKPLPRH PNETYEDYGK GLPKKLERRD EEHDIQNDDN | 60 |
| QIPQEDGKQS SHDNQSPAES NTGSTTPVDP SEGDEDSNKE YLNDLQFKKK DLKKPRTQSF | 120 |
| QLVLSTASLK SLRQTEPANP PALQRNSSNL GNMSLTGTGN HKNFQSFIQA PVLSSVSNLR | 180 |
| ALDNIVIGQQ LPFDHKDPPS ESELNRLRHN SNVTQSSTQP VIKDDRDDDD EYREETILQQ | 240 |
| QKLTLNALKK LSLSLAPIIR SDEETGPESR QLTTKLLNGL LKPESKLEQR KEEGNNKLQT | 300 |
| ENRTKPYQPA QVDLSLFASL TRQNKHLTEP QLPPSQGKPT NQPNERSKNA PLSSERDYHI | 360 |
| DMQRQMLSMQ NQTNGSEPAQ KLYGQTLLIE PDPVTDIKSA PQQTKGPINH SNLNLHSHKL | 420 |
| QQSQSQVLMN APSQPLSHSQ SLGQLQSQKS ETELNPVVPP MDMNVRNRKT SQAEDTLPKH | 480 |
| ASLADKRIQQ IKGFRNPMYI PAVLRKTLDK DADLGPLSSN LTDTGRESYF EGLSRVGLPN | 540 |
| RDHGSSSNSI RSVDLADNTI ASPSLPFTGP YTLNKKQYEH ILKAAPTRKH WLKDEAVSEC | 600 |
| GITTCRKNFN FFERRHHCRR CGGIFCKEHT LHYLYINHLA QFTTGGRGTL SRVCDNCIEE | 660 |
| YNEFMKQEFG VTCHRPRSSI DVTTAPLEEP EVKPSNKPAL SPRKELLKYK NPHQRPRNQI | 720 |
| SPQVQIGKGY AREDQSTEQV AGSVPANWSV KIDTVRLLLR TVIFISIAGA AISSRLFSVI | 780 |
| RFESIIHEFD PWFNFRATKY LVSHSFYDFL NWFDDRTWYP LGRVTGGTLY PGLMVTSGVI | 840 |
| WHTLRDWFGL PVDIRNICVM LAPAFSGITA IFTYYFTKEM KDSNAGLLAA IFMGIAPGYI | 900 |
| SRSVAGSYDN EAIAITLLMA TFYLWIKAMK VGSIFYGTLT ALSYFYMVSA WGGYVFITNL | 960 |
| IPLHVFVLIL MGRYNHRLYT AYTTWYALGT IASMQIPFVG FLPIRSNDHM AALGVFGLLQ | 1020 |
| LVAFGAYVRS NVPTAQFKVF LTVSLFFTTL LGVAGLTALT AAGWIAPWTG RFYSLWDTNY | 1080 |
| AKIHIPIIAS VSEHQPTAWP AFFFDTNMLI WLFPAGVFFC FQEMRDEHLF IIIYGVLGSY | 1140 |
| FAGVMVRLML TLTPVVCVAA AIALSKLFDV YMDVSDIFGS EAAVEEVSEK KKKGPAKQGK | 1200 |
| QFPIFKLLSK GVVLASFVYY LVFFVLHCTW VTSHAYSSPS VVLASRNPDG TQNIIDDYRE | 1260 |
| AYYWLRMNTP EDAKVMAWWD YGYQIGGMAD RTTLVDNNTW NNTHIATVGK AMATNEENAY | 1320 |
| EILKKHDVDY VLVIFGGVLG YSGDDMNKFL WMVRISEGIW PDEIHERDYF STRGEYKVDK | 1380 |
| DASETMKNSM MYKMSYYRFA ELFGGREAQD RARGQVIPSD PITLDTVEEA FTSQNWIVRI | 1440 |
| YKVKDLDNLG REPKAAADFE RGTSGANKRV RAVKKPALDL RV | 1482 |
Enzyme Prediction help
| EC | 2.4.99.18:21 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT66 | 757 | 1402 | 3.4e-192 | 0.8571428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396873 | STT3 | 0.0 | 758 | 1237 | 1 | 478 | Oligosaccharyl transferase STT3 subunit. This family consists of the oligosaccharyl transferase STT3 subunit and related proteins. The STT3 subunit is part of the oligosaccharyl transferase (OTase) complex of proteins and is required for its activity. In eukaryotes, OTase transfers a lipid-linked core-oligosaccharide to selected asparagine residues in the ER. In the archaea STT3 occurs alone, rather than in an OTase complex, and is required for N-glycosylation of asparagines. |
| 224206 | Stt3 | 5.92e-151 | 756 | 1476 | 16 | 761 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
| 277299 | FYVE_scVPS27p_like | 1.21e-25 | 590 | 657 | 1 | 58 | FYVE domain found in Saccharomyces cerevisiae vacuolar protein sorting-associated protein 27 (scVps27p) and similar proteins. scVps27p, also termed Golgi retention defective protein 11, is the putative yeast counterpart of the mammalian protein Hrs and is involved in endosome maturation. It is a mono-ubiquitin-binding protein that interacts with ubiquitinated cargoes, such as Hse1p, and is required for protein sorting into the multivesicular body. Vps27p forms a complex with Hse1p. The complex binds ubiquitin and mediates endosomal protein sorting. At the endosome, Vps27p and a trimeric protein complex, ESCRT-1, bind ubiquitin and are important for multivesicular body (MVB) sorting. Vps27p contains an N-terminal VHS (Vps27/Hrs/STAM) domain, a FYVE domain that binds PtdIns3P, followed by two ubiquitin-interacting motifs (UIMs), and a C-terminal clathrin-binding motif. |
| 214499 | FYVE | 5.36e-22 | 590 | 662 | 3 | 67 | Protein present in Fab1, YOTB, Vac1, and EEA1. The FYVE zinc finger is named after four proteins where it was first found: Fab1, YOTB/ZK632.12, Vac1, and EEA1. The FYVE finger has been shown to bind two Zn2+ ions. The FYVE finger has eight potential zinc coordinating cysteine positions. The FYVE finger is structurally related to the PHD finger and the RING finger. Many members of this family also include two histidines in a motif R+HHC+XCG, where + represents a charged residue and X any residue. The FYVE finger functions in the membrane recruitment of cytosolic proteins by binding to phosphatidylinositol 3-phosphate (PI3P), which is prominent on endosomes. The R+HHC+XCG motif is critical for PI3P binding. |
| 396091 | FYVE | 2.19e-21 | 590 | 663 | 2 | 68 | FYVE zinc finger. The FYVE zinc finger is named after four proteins that it has been found in: Fab1, YOTB/ZK632.12, Vac1, and EEA1. The FYVE finger has been shown to bind two Zn++ ions. The FYVE finger has eight potential zinc coordinating cysteine positions. Many members of this family also include two histidines in a motif R+HHC+XCG, where + represents a charged residue and X any residue. We have included members which do not conserve these histidine residues but are clearly related. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWU87245.1|GT66 | 0.0 | 1 | 1482 | 1 | 1497 |
| QRG38406.1|GT66 | 0.0 | 750 | 1481 | 10 | 742 |
| QEL60034.1|GT66 | 0.0 | 750 | 1481 | 10 | 742 |
| QEO20766.1|GT66 | 0.0 | 750 | 1481 | 10 | 742 |
| QWW23032.1|GT66 | 0.0 | 750 | 1472 | 10 | 733 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6C26_A | 0.0 | 758 | 1482 | 15 | 718 | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex [Saccharomyces cerevisiae S288C],6EZN_F Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex [Saccharomyces cerevisiae S288C],7OCI_F Chain F, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 [Saccharomyces cerevisiae S288C] |
| 6S7T_A | 2.32e-277 | 743 | 1459 | 63 | 791 | Cryo-EM structure of human oligosaccharyltransferase complex OST-B [Homo sapiens] |
| 6FTG_5 | 6.23e-267 | 758 | 1450 | 17 | 701 | Subtomogram average of OST-containing ribosome-translocon complexes from canine rough microsomal membranes [Canis lupus familiaris],6FTI_5 Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome [Canis lupus familiaris] |
| 6S7O_A | 6.94e-266 | 758 | 1450 | 17 | 701 | Cryo-EM structure of human oligosaccharyltransferase complex OST-A [Homo sapiens] |
| 6FTJ_5 | 6.26e-264 | 758 | 1445 | 17 | 696 | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome [Canis lupus familiaris] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P39007|STT3_YEAST | 0.0 | 758 | 1482 | 15 | 718 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=STT3 PE=1 SV=2 |
| sp|O94335|STT3_SCHPO | 3.98e-312 | 756 | 1482 | 21 | 752 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit stt3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=stt3 PE=3 SV=1 |
| sp|Q3TDQ1|STT3B_MOUSE | 2.71e-277 | 743 | 1454 | 60 | 783 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Mus musculus OX=10090 GN=Stt3b PE=1 SV=2 |
| sp|E2RG47|STT3B_CANLF | 8.45e-277 | 743 | 1459 | 63 | 791 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Canis lupus familiaris OX=9615 GN=STT3B PE=1 SV=1 |
| sp|Q8TCJ2|STT3B_HUMAN | 1.19e-276 | 743 | 1459 | 63 | 791 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Homo sapiens OX=9606 GN=STT3B PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000004 |
TMHMM Annotations download full data without filtering help
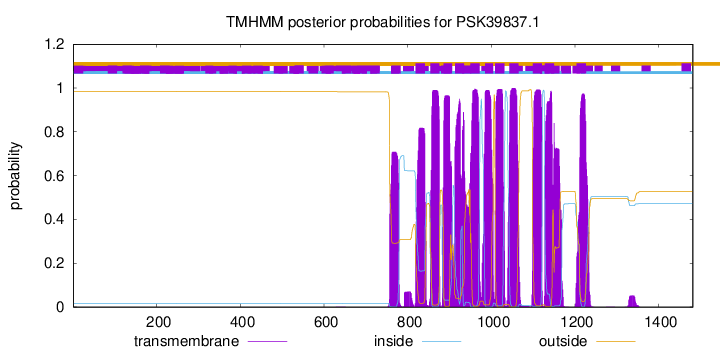
| Start | End |
|---|---|
| 819 | 841 |
| 854 | 876 |
| 886 | 906 |
| 913 | 935 |
| 950 | 972 |
| 985 | 1004 |
| 1009 | 1031 |
| 1043 | 1065 |
| 1099 | 1121 |
| 1128 | 1150 |
| 1204 | 1226 |
