You are browsing environment: FUNGIDB
CAZyme Information: PSK39621.1
You are here: Home > Sequence: PSK39621.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK39621.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.258:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT58 | 46 | 438 | 2.3e-129 | 0.9945054945054945 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398745 | ALG3 | 8.51e-166 | 46 | 438 | 1 | 358 | ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 471 | 619 | 1089 | |
| 3.27e-273 | 12 | 470 | 22 | 480 | |
| 1.33e-272 | 12 | 470 | 22 | 480 | |
| 1.05e-271 | 12 | 472 | 22 | 482 | |
| 3.00e-271 | 12 | 460 | 22 | 470 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-198 | 4 | 460 | 3 | 460 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=ALG3 PE=3 SV=1 |
|
| 2.04e-192 | 8 | 472 | 14 | 476 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Scheffersomyces stipitis (strain ATCC 58785 / CBS 6054 / NBRC 10063 / NRRL Y-11545) OX=322104 GN=ALG3 PE=3 SV=2 |
|
| 1.37e-187 | 3 | 460 | 17 | 473 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Meyerozyma guilliermondii (strain ATCC 6260 / CBS 566 / DSM 6381 / JCM 1539 / NBRC 10279 / NRRL Y-324) OX=294746 GN=ALG3 PE=3 SV=2 |
|
| 4.66e-135 | 11 | 471 | 5 | 464 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=ALG3 PE=3 SV=2 |
|
| 1.73e-90 | 26 | 456 | 34 | 450 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=ALG3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000043 | 0.000029 |
TMHMM Annotations download full data without filtering help
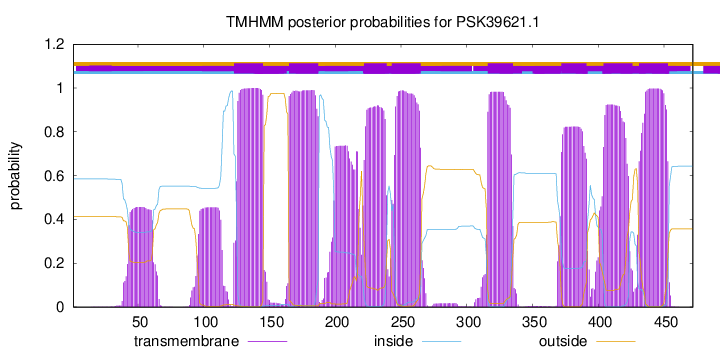
| Start | End |
|---|---|
| 123 | 145 |
| 165 | 187 |
| 222 | 239 |
| 243 | 265 |
| 316 | 335 |
| 372 | 391 |
| 404 | 426 |
| 431 | 453 |
