You are browsing environment: FUNGIDB
CAZyme Information: PSK38751.1
You are here: Home > Sequence: PSK38751.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK38751.1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 572736; End:574865 Strand: - | |||||||||||
Full Sequence Download help
| MKITTEQLFI LGVLLRIGFF LFGLFQDKYM AVRYTDIDYV VFSDAAKYVA DDKSPYQRET | 60 |
| YRYTPMLAWI LLPVTFGGNW VHYGKALFML CDIITGALIT EIVKKETPGS STKDTFFQRN | 120 |
| KTTILSAIWL LNPMVITIST RGSSESVLSC FIMLAVANLF RDQYIMSALF LGLSIHFKIY | 180 |
| PIIYLPSIML FLASKKPLLI KAWQNIPFLG WVNTANLSYL VATLVSFAVP TYLMYDFYGY | 240 |
| EFLYHSYLYH LTRLDHRHNF SLYNLALYLK SAQDYLPQNL DSENFLTIAL QSIEKAAFAP | 300 |
| QIVLSGLVIP LVLARRNLTA CLFIQTLTFV TFNKVMTSQY FIWFLIFLPS YLATSQLLSK | 360 |
| LNARKGSLML LLWIASQASM SRMELYSPEG LRIDGRRWNE LRRFECQINT HPHSSDGSSY | 420 |
| VEHGNTKVMC IVKGPMEPRT RAQQDQDNAT LDININVASF STLERKKRSK NEKRLIELKT | 480 |
| TLERTFEKSV LTHLYPKTLI EISVQVLAQD GGMLATITNA ITLALIDAGI SIYDYVSAVT | 540 |
| VGLHDQTPLL DLNTLEEGDV SNLTVGVVGK SEKLAMLLLE DKMPLDHLES VLGIAIAGSH | 600 |
| KVRELLDEEQ SYKVKFKV | 618 |
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 138 | 378 | 7.6e-72 | 0.8473282442748091 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 206775 | RNase_PH_RRP41 | 5.34e-122 | 390 | 609 | 1 | 220 | RRP41 subunit of eukaryotic exosome. The RRP41 subunit of eukaryotic exosome is a member of the RNase_PH family, named after the bacterial Ribonuclease PH, a 3'-5' exoribonuclease. Structurally all members of this family form hexameric rings (trimers of Rrp41-Rrp45, Rrp46-Rrp43, and Mtr3-Rrp42 dimers). The eukaryotic exosome core is composed of six individually encoded RNase PH-like subunits and three additional proteins (Rrp4, Csl4 and Rrp40) that form a stable cap and contain RNA-binding domains. The RNase PH-like subunits are no longer phosphorolytic enzymes, the exosome directly associates with Rrp44 and Rrp6, hydrolytic exoribonucleases related to bacterial RNase II/R and RNase D. The exosome plays an important role in RNA turnover. It plays a crucial role in the maturation of stable RNA species such as rRNA, snRNA and snoRNA, quality control of mRNA, and the degradation of RNA processing by-products and non-coding transcripts. |
| 178434 | PLN02841 | 1.67e-72 | 3 | 377 | 6 | 375 | GPI mannosyltransferase |
| 223761 | Rph | 5.25e-67 | 384 | 608 | 1 | 227 | Ribonuclease PH [Translation, ribosomal structure and biogenesis]. |
| 252941 | Mannosyl_trans | 9.10e-66 | 138 | 378 | 1 | 220 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 235187 | PRK03983 | 2.12e-41 | 389 | 605 | 12 | 227 | exosome complex exonuclease Rrp41; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWU88835.1|GT50 | 0.0 | 1 | 609 | 1 | 609 |
| QWW23894.1|GT50 | 1.28e-302 | 3 | 609 | 4 | 624 |
| QRG39151.1|GT50 | 4.04e-160 | 3 | 377 | 4 | 379 |
| QEO20435.1|GT50 | 3.41e-156 | 3 | 377 | 4 | 379 |
| QEL61505.1|GT50 | 1.37e-155 | 3 | 377 | 4 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5G06_B | 1.26e-98 | 380 | 609 | 1 | 231 | Cryo-EM structure of yeast cytoplasmic exosome [Saccharomyces cerevisiae],6LQS_R1 Chain R1, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7AJT_EC Chain EC, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7AJU_EC Chain EC, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C],7D4I_R1 Chain R1, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C] |
| 4IFD_B | 1.34e-98 | 380 | 609 | 3 | 233 | Crystal structure of an 11-subunit eukaryotic exosome complex bound to RNA [Saccharomyces cerevisiae S288C],5C0W_B Structure of a 12-subunit nuclear exosome complex bound to single-stranded RNA substrates [Saccharomyces cerevisiae S288C],5C0X_B Structure of a 12-subunit nuclear exosome complex bound to structured RNA [Saccharomyces cerevisiae S288C],6FSZ_BB Chain BB, Exosome complex component SKI6 [Saccharomyces cerevisiae S288C] |
| 5JEA_B | 1.39e-98 | 380 | 609 | 4 | 234 | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA [Saccharomyces cerevisiae S288C],5OKZ_B Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_L Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_V Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C],5OKZ_f Crystal Strucrure of the Mpp6 Exosome complex [Saccharomyces cerevisiae S288C] |
| 4OO1_B | 1.44e-98 | 380 | 609 | 5 | 235 | Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA [Saccharomyces cerevisiae S288C],5K36_B Structure of an eleven component nuclear RNA exosome complex bound to RNA [Saccharomyces cerevisiae S288C],5VZJ_B STRUCTURE OF A TWELVE COMPONENT MPP6-NUCLEAR RNA EXOSOME COMPLEX BOUND TO RNA [Saccharomyces cerevisiae S288C] |
| 2WP8_B | 2.50e-98 | 380 | 609 | 1 | 231 | yeast rrp44 nuclease [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q6BHI9|GPI14_DEBHA | 2.44e-126 | 1 | 379 | 2 | 368 | GPI mannosyltransferase 1 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI14 PE=3 SV=2 |
| sp|Q5AMR5|GPI14_CANAL | 1.44e-114 | 6 | 379 | 5 | 356 | GPI mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI14 PE=3 SV=1 |
| sp|P46948|RRP41_YEAST | 6.46e-98 | 380 | 609 | 1 | 231 | Exosome complex component SKI6 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SKI6 PE=1 SV=1 |
| sp|Q6CRE7|GPI14_KLULA | 1.52e-87 | 12 | 378 | 12 | 344 | GPI mannosyltransferase 1 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=GPI14 PE=3 SV=1 |
| sp|Q6FXQ5|GPI14_CANGA | 8.98e-84 | 2 | 378 | 3 | 353 | GPI mannosyltransferase 1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=GPI14 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000012 | 0.000013 |
TMHMM Annotations download full data without filtering help
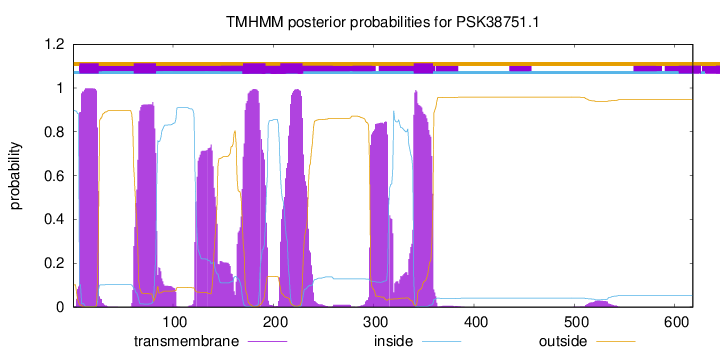
| Start | End |
|---|---|
| 7 | 26 |
| 61 | 83 |
| 170 | 192 |
| 207 | 229 |
| 340 | 359 |
