You are browsing environment: FUNGIDB
CAZyme Information: PSK36426.1
You are here: Home > Sequence: PSK36426.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
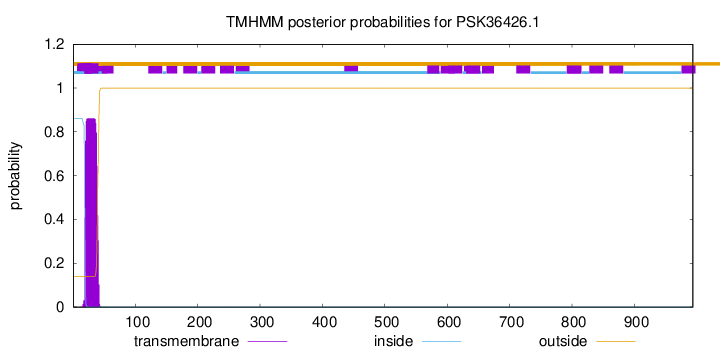
TMHMM annotations
Basic Information help
| Species | [Candida] pseudohaemulonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] pseudohaemulonis | |||||||||||
| CAZyme ID | PSK36426.1 | |||||||||||
| CAZy Family | GH132 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA2 | 104 | 339 | 2.3e-47 | 0.9098039215686274 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173825 | ascorbate_peroxidase | 1.39e-118 | 70 | 343 | 2 | 253 | Ascorbate peroxidases and cytochrome C peroxidases. Ascorbate peroxidases are a subgroup of heme-dependent peroxidases of the plant superfamily that share a heme prosthetic group and catalyze a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. Along with related catalase-peroxidases, ascorbate peroxidases belong to class I of the plant superfamily. Ascorbate peroxidases are found in the chloroplasts and/or cytosol of algae and plants, where they have been shown to control the concentration of lethal hydrogen peroxide molecules. The yeast cytochrome c peroxidase is a divergent member of the family; it forms a complex with cytochrome c to catalyze the reduction of hydrogen peroxide to water. |
| 270250 | PBP2_PDT_like | 7.12e-66 | 547 | 708 | 30 | 184 | Catalytic domain of prephenate dehydratase and similar proteins; the type 2 periplasmic binding protein fold. Prephenate dehydratase (PDT, EC:4.2.1.51) converts prephenate to phenylpyruvate through dehydration and decarboxylation reactions. PDT plays a key role in the biosynthesis of L-Phe in organisms that utilize the shikimate pathway. PDT is allosterically regulated by L-Phe and other amino acids. The catalytic PDT domain consists of two similar subdomains with a cleft in between, which hosts the highly conserved active site. In gram-postive bacteria and archaea, PDT is a monofunctional enzyme, consisting of a catalytic domain (PDT domain) and a regulatory domain (ACT) (aspartokinase, chorismate mustase domain). In gram-negative bacteria, PDT exists as fusion protein with chorismate mutase (CM), forming a bifunctional enzyme, P-protein (PheA). The CM in the P-protein catalyzes the pericycle isomerization of chorismate to prephenate that serves as a substrate for PDT. The CM and PDT are essentail enzymes for the biosynthesis of aromatic amino acids in microorganisms but are not found in humans. Thus, both CM and PDT can potentially serve as drug targets against microbial pathogens. The PDT domain has the same structural fold as the type 2 periplasmic binding proteins (PBP2), many of which are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
| 240234 | PTZ00027 | 5.70e-65 | 359 | 541 | 1 | 182 | 60S ribosomal protein L6; Provisional |
| 223155 | PheA2 | 8.17e-63 | 546 | 769 | 30 | 233 | Prephenate dehydratase [Amino acid transport and metabolism]. |
| 178218 | PLN02608 | 5.15e-57 | 107 | 339 | 33 | 243 | L-ascorbate peroxidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.51e-211 | 1 | 358 | 1278 | 1635 | |
| 5.72e-204 | 56 | 358 | 54 | 356 | |
| 5.72e-204 | 56 | 358 | 54 | 356 | |
| 1.62e-203 | 56 | 358 | 54 | 356 | |
| 1.65e-143 | 76 | 358 | 94 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.13e-99 | 359 | 548 | 1 | 190 | Chain q, Ribosomal 60S subunit protein L9B [Candida albicans SC5314],7Q0F_q Chain q, Ribosomal 60S subunit protein L9B [Candida albicans SC5314],7Q0P_q Chain q, Ribosomal 60S subunit protein L9B [Candida albicans SC5314] |
|
| 5.06e-95 | 65 | 359 | 3 | 294 | Structure of isoniazid (INH) bound to cytochrome c peroxidase mutant N184R Y36A [Saccharomyces cerevisiae] |
|
| 7.55e-95 | 69 | 359 | 9 | 296 | Structure of cytochrome c peroxidase mutant N184R Y36A [Saccharomyces cerevisiae],4A6Z_A Cytochrome c peroxidase with bound guaiacol [Saccharomyces cerevisiae] |
|
| 1.05e-94 | 69 | 359 | 9 | 296 | cytochrome c peroxidase in complex with phenol [Saccharomyces cerevisiae] |
|
| 2.42e-94 | 69 | 359 | 4 | 291 | Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M25_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M26_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M27_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M28_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M29_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2A_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2B_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2C_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2D_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2E_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2F_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2G_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2H_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae],3M2I_A Chain A, Cytochrome c peroxidase, mitochondrial [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.71e-137 | 55 | 358 | 59 | 359 | Cytochrome c peroxidase, mitochondrial OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=CCP1 PE=3 SV=1 |
|
| 5.74e-129 | 62 | 358 | 71 | 364 | Cytochrome c peroxidase, mitochondrial OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CCP1 PE=3 SV=2 |
|
| 1.56e-95 | 63 | 352 | 52 | 336 | Cytochrome c peroxidase, mitochondrial OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=CCP1 PE=3 SV=1 |
|
| 1.08e-92 | 62 | 359 | 65 | 357 | Cytochrome c peroxidase, mitochondrial OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=CAGL0K08184g PE=3 SV=1 |
|
| 1.71e-92 | 62 | 359 | 66 | 361 | Cytochrome c peroxidase, mitochondrial OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CCP1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999832 | 0.000195 |