You are browsing environment: FUNGIDB
CAZyme Information: PPTG_19662-t26_1-p1
You are here: Home > Sequence: PPTG_19662-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_19662-t26_1-p1 | |||||||||||
| CAZy Family | PL4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 9.79e-21 | 69 | 206 | 1 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 4.85e-17 | 62 | 210 | 25 | 174 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 380914 | SET | 1.71e-06 | 208 | 263 | 21 | 72 | SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain superfamily. The Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domain superfamily corresponds to SET domain-containing lysine methyltransferases, which catalyze site and state-specific methylation of lysine residues in histones that are fundamental in epigenetic regulation of gene activation and silencing in eukaryotic organisms. SET domains appear to be protein-protein interaction domains. It has been demonstrated that SET domains mediate interactions with a family of proteins that display similarity with dual-specificity phosphatases (dsPTPases). A subset of SET domains has been called PR domains. These domains are divergent in sequence from other SET domains, but also appear to mediate protein-protein interaction. The SET domain consists of two regions known as N-SET and C-SET. C-SET forms an unusual and conserved knot-like structure of probable functional importance. In addition to N-SET and C-SET, an insert region (I-SET) and flanking regions of high structural variability form part of the overall structure. Some family members contain a pre-SET domain, which is found in a number of histone methyltransferases (HMTase), and a post-SET domain, which harbors a zinc-binding site. |
| 178402 | PLN02805 | 9.86e-06 | 59 | 203 | 122 | 268 | D-lactate dehydrogenase [cytochrome] |
| 380920 | SET_LegAS4-like | 1.58e-05 | 199 | 263 | 59 | 117 | SET domain found in Legionella pneumophila type IV secretion system effector LegAS4 and similar proteins. LegAS4 is a type IV secretion system effector of Legionella pneumophila. It contains a SET domain that is involved in the modification of Lys4 of histone H3 (H3K4) in the nucleolus of the host cell, thereby enhancing heterochromatic rDNA transcription. It also contains an ankyrin repeat domain of unknown function at its C-terminal region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.09e-110 | 1 | 221 | 1 | 220 | |
| 4.22e-98 | 1 | 221 | 1 | 251 | |
| 2.50e-33 | 27 | 232 | 21 | 233 | |
| 3.46e-33 | 27 | 232 | 21 | 233 | |
| 6.61e-33 | 27 | 232 | 21 | 233 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.51e-35 | 27 | 232 | 1 | 213 | Chain A, FAD-binding PCMH-type domain-containing protein [Fusarium graminearum PH-1],6YJI_B Chain B, FAD-binding PCMH-type domain-containing protein [Fusarium graminearum PH-1] |
|
| 4.71e-30 | 32 | 237 | 24 | 240 | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],5L6F_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylobiose [Thermothelomyces thermophilus ATCC 42464],5L6G_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylose [Thermothelomyces thermophilus ATCC 42464] |
|
| 3.21e-27 | 38 | 254 | 17 | 241 | Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens],6EO5_B Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens] |
|
| 3.21e-27 | 38 | 254 | 17 | 241 | Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens],6EO4_B Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens] |
|
| 5.76e-27 | 37 | 222 | 5 | 194 | Crystal structure of carbohydrate oxidase from Microdochium nivale [Microdochium nivale],3RJA_A Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue [Microdochium nivale] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.42e-29 | 32 | 237 | 24 | 240 | Xylooligosaccharide oxidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=xylO PE=1 SV=1 |
|
| 5.77e-27 | 37 | 222 | 31 | 218 | FAD-linked oxidoreductase subF OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=subF PE=3 SV=1 |
|
| 9.86e-27 | 31 | 222 | 21 | 216 | Carbohydrate oxidase OS=Microdochium nivale OX=5520 GN=MnCO PE=1 SV=2 |
|
| 1.82e-25 | 37 | 222 | 31 | 218 | FAD-linked oxidoreductase dpmaF OS=Metarhizium anisopliae OX=5530 GN=dpmaF PE=1 SV=1 |
|
| 1.08e-24 | 35 | 215 | 27 | 207 | Chitooligosaccharide oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=chitO PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
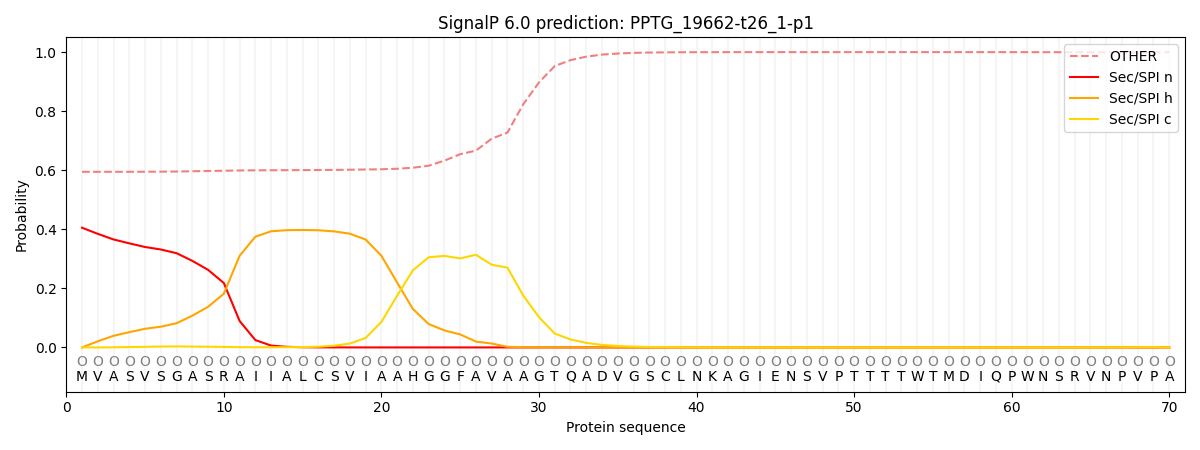
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.610560 | 0.389434 |
