You are browsing environment: FUNGIDB
CAZyme Information: PPTG_18899-t26_1-p1
Basic Information
help
| Species |
Phytophthora parasitica
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica
|
| CAZyme ID |
PPTG_18899-t26_1-p1
|
| CAZy Family |
GT71 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 979 |
KI669649|CGC1 |
105560.35 |
6.7302 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PparasiticaINRA-310 |
23240 |
761204 |
119 |
23121
|
|
| Gene Location |
No EC number prediction in PPTG_18899-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
467 |
712 |
1.2e-20 |
0.45351043643263755 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 404168
|
Beta_helix |
5.06e-06 |
597 |
717 |
3 |
117 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| 404168
|
Beta_helix |
0.005 |
595 |
651 |
93 |
149 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.67e-13 |
435 |
710 |
148 |
379 |
| 4.00e-13 |
407 |
712 |
191 |
454 |
| 3.57e-12 |
407 |
712 |
187 |
450 |
| 8.20e-12 |
409 |
711 |
190 |
450 |
| 1.42e-11 |
295 |
712 |
76 |
447 |
PPTG_18899-t26_1-p1 has no PDB hit.
PPTG_18899-t26_1-p1 has no Swissprot hit.
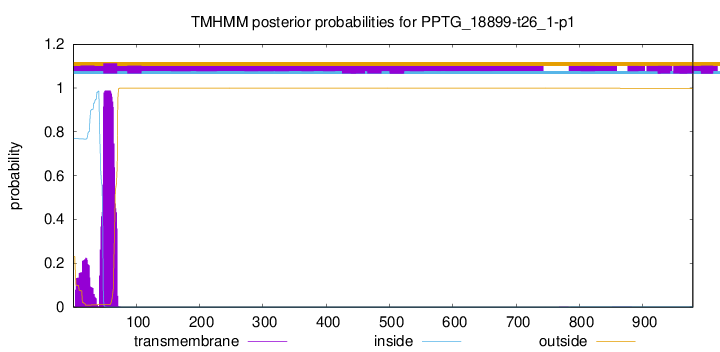
This protein is predicted as OTHER
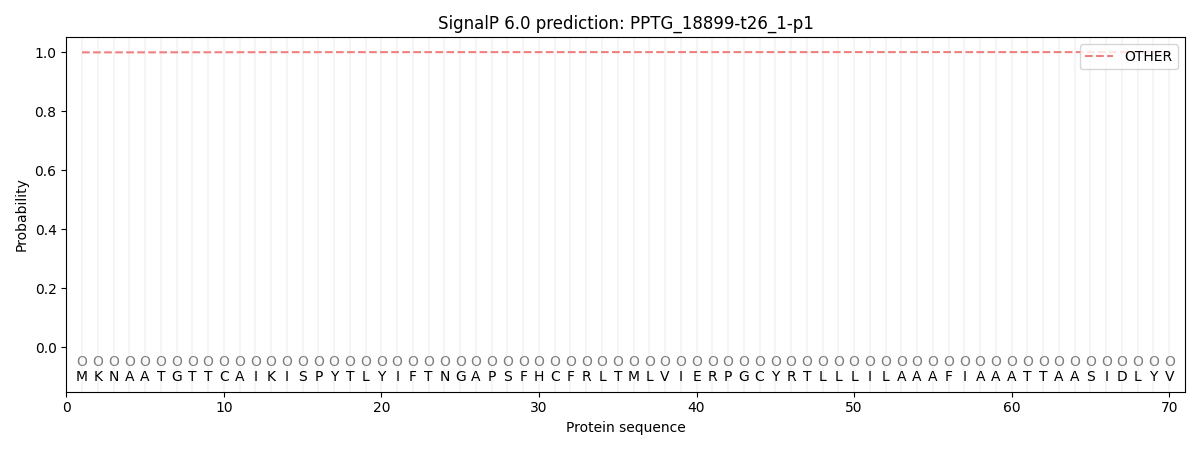
| Other |
SP_Sec_SPI |
CS Position |
| 0.999311 |
0.000688 |
|