You are browsing environment: FUNGIDB
CAZyme Information: PPTG_18120-t26_1-p1
You are here: Home > Sequence: PPTG_18120-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_18120-t26_1-p1 | |||||||||||
| CAZy Family | GT59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.164:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 37 | 490 | 6.4e-134 | 0.9846491228070176 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 405300 | Glyco_hydr_30_2 | 6.53e-17 | 29 | 239 | 3 | 195 | O-Glycosyl hydrolase family 30. |
| 227807 | XynC | 2.62e-07 | 135 | 490 | 89 | 426 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.80e-229 | 3 | 493 | 4 | 497 | |
| 3.10e-164 | 15 | 496 | 21 | 498 | |
| 3.89e-160 | 9 | 496 | 16 | 499 | |
| 4.45e-159 | 9 | 496 | 16 | 499 | |
| 4.45e-159 | 9 | 496 | 16 | 499 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-128 | 5 | 497 | 2 | 478 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
|
| 1.25e-16 | 5 | 367 | 6 | 445 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
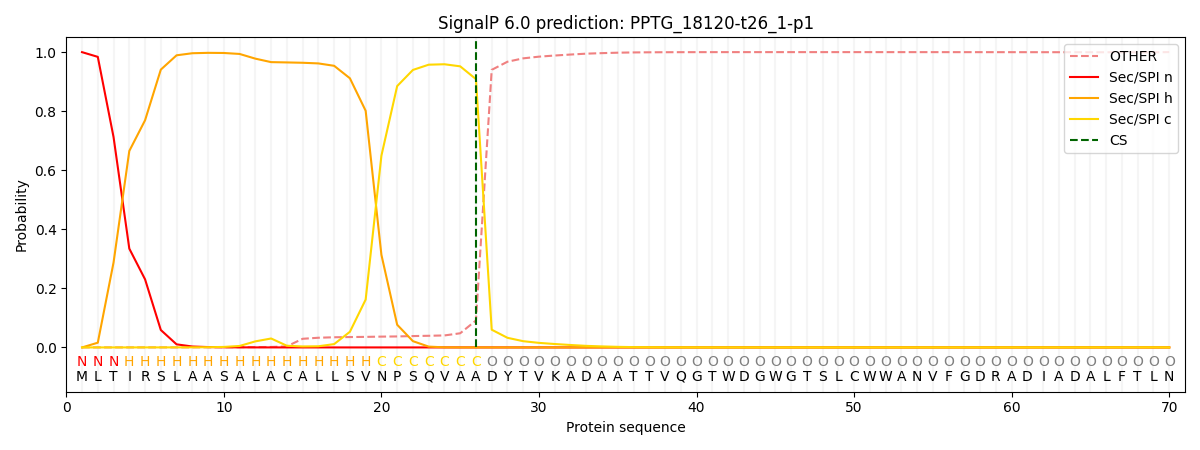
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001038 | 0.998925 | CS pos: 26-27. Pr: 0.9085 |
